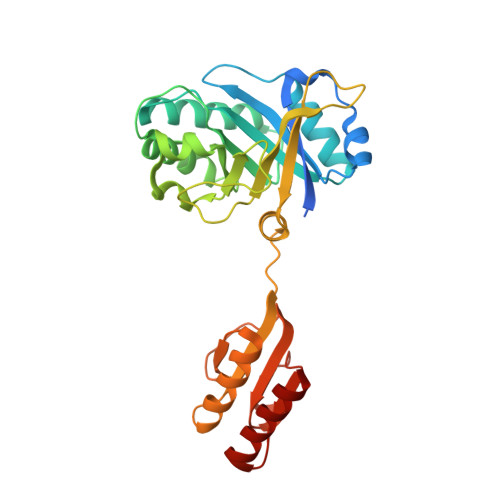
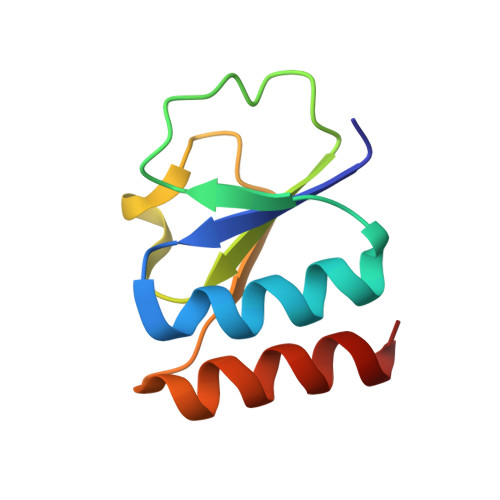
Crystal Structure of the Ctp1L Endolysin Reveals How its Activity is Regulated by a Secondary Translation Product.
Dunne, M., Leicht, S., Krichel, B., Mertens, H.D.T., Thompson, A., Krijgsveld, J., Svergun, D.I., Gomez-Torres, N., Garde, S., Uetrecht, C., Narbad, A., Mayer, M.J., Meijers, R.(2016) J Biol Chem 291: 4882
- PubMed: 26683375
- DOI: https://doi.org/10.1074/jbc.M115.671172
- Primary Citation of Related Structures:
5A6S - PubMed Abstract:
Bacteriophages produce endolysins, which lyse the bacterial host cell to release newly produced virions. The timing of lysis is regulated and is thought to involve the activation of a molecular switch. We present a crystal structure of the activated endolysin CTP1L that targets Clostridium tyrobutyricum, consisting of a complex between the full-length protein and an N-terminally truncated C-terminal cell wall binding domain (CBD). The truncated CBD is produced through an internal translation start site within the endolysin gene. Mutants affecting the internal translation site change the oligomeric state of the endolysin and reduce lytic activity. The activity can be modulated by reconstitution of the full-length endolysin-CBD complex with free CBD. The same oligomerization mechanism applies to the CD27L endolysin that targets Clostridium difficile and the CS74L endolysin that targets Clostridium sporogenes. When the CTP1L endolysin gene is introduced into the commensal bacterium Lactococcus lactis, the truncated CBD is also produced, showing that the alternative start codon can be used in other bacterial species. The identification of a translational switch affecting oligomerization presented here has implications for the design of effective endolysins for the treatment of bacterial infections.
Organizational Affiliation:
From the European Molecular Biology Laboratory, Notkestrasse 85, 22607 Hamburg, Germany.