The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses.
Dimaio, F., Chen, C., Yu, X., Frenz, B., Hsu, Y., Lin, N., Egelman, E.H.(2015) Nat Struct Mol Biol 22: 642
- PubMed: 26167882
- DOI: https://doi.org/10.1038/nsmb.3054
- Primary Citation of Related Structures:
5A2T - PubMed Abstract:
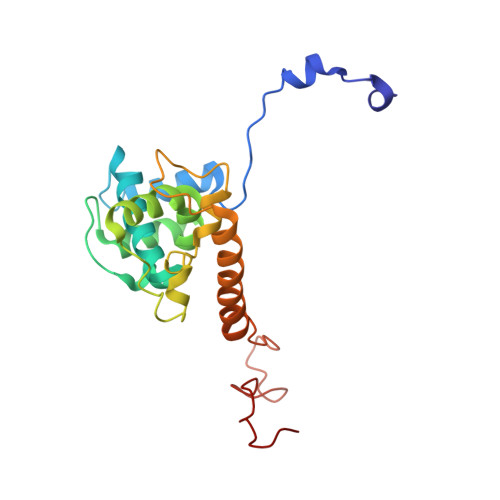
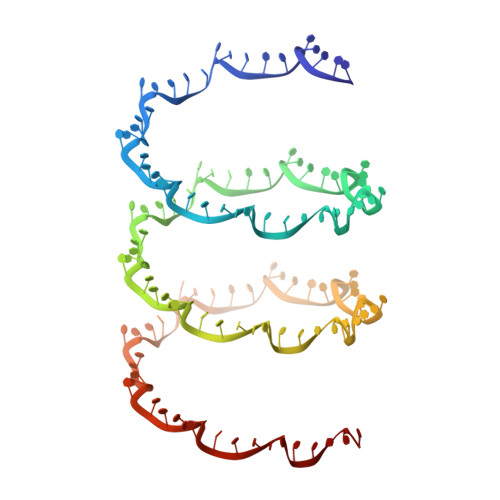
Flexible filamentous plant viruses cause more than half the viral crop damage in the world but are also potentially useful for biotechnology. Structural studies began more than 75 years ago but have failed, owing to the virion's extreme flexibility. We have used cryo-EM to generate an atomic model for bamboo mosaic virus, which reveals flexible N- and C-terminal extensions that allow deformation while still maintaining structural integrity.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, Washington, USA.