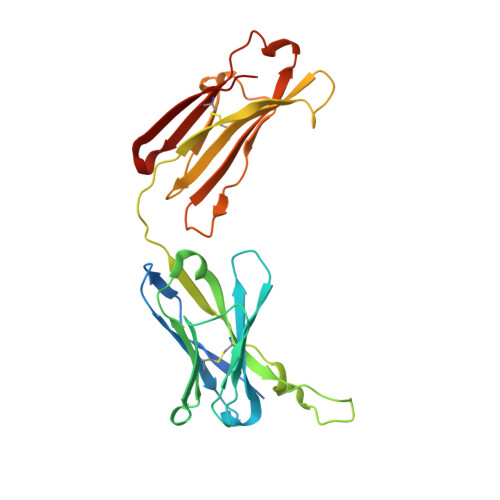
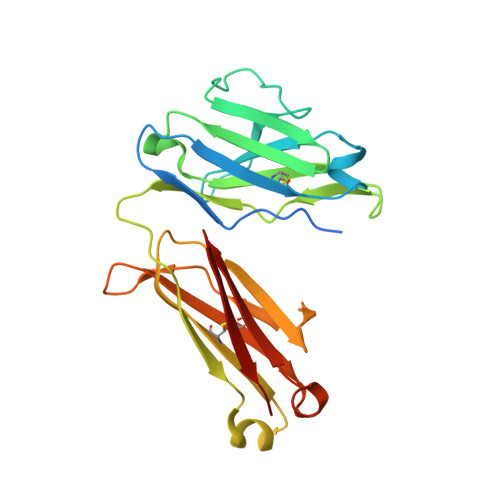
Crystal structure of the Fab region of a neutralizing antibody against granulocyte-macrophage colony-stimulating factor.
Angkawidjaja, C., Torashima, T.(2019) Acta Crystallogr F Struct Biol Commun 75: 634-639
- PubMed: 31584011
- DOI: https://doi.org/10.1107/S2053230X1901238X
- Primary Citation of Related Structures:
5ZMJ - PubMed Abstract:
An increased level of granulocyte-macrophage colony-stimulating factor has a potential role in the development of autoimmune diseases, and the neutralization of its activity by monoclonal antibodies is a promising therapy for some diseases. Here, the crystal structure of the Fab region of EV1007, a fully human antibody expressed in Chinese hamster ovary cells that was developed from human peripheral blood mononuclear cells, is described. The structure closely resembles that of MB007, which is the Fab region of the same antibody expressed in Escherichia coli [Blech et al. (2012), Biochem. J. 447, 205-215], except at the hinge regions between the immunoglobulin domains and the H3 loop region. This paper presents evidence for the flexibility of the hinge and H3 loop regions of the antibody based on the comparison of two independently solved crystal structures.
Organizational Affiliation:
Antibody Drug Discovery Laboratory, Evec Inc., Technopark 1-chome, Sapporo Electronics Center, Sapporo, Hokkaido, Japan.