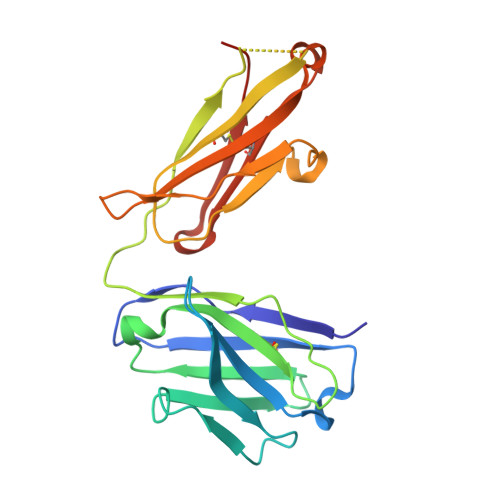
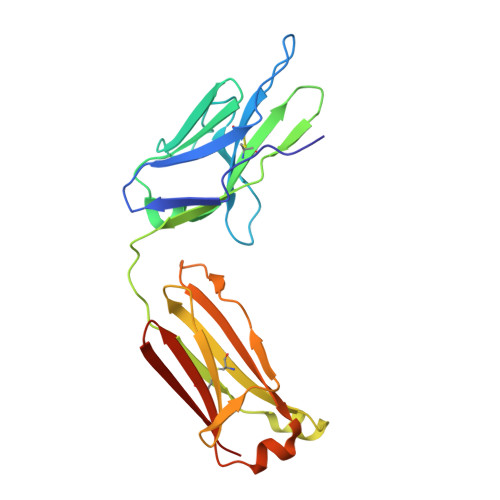
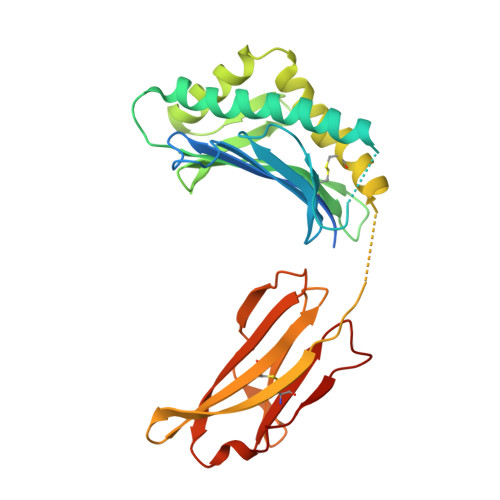
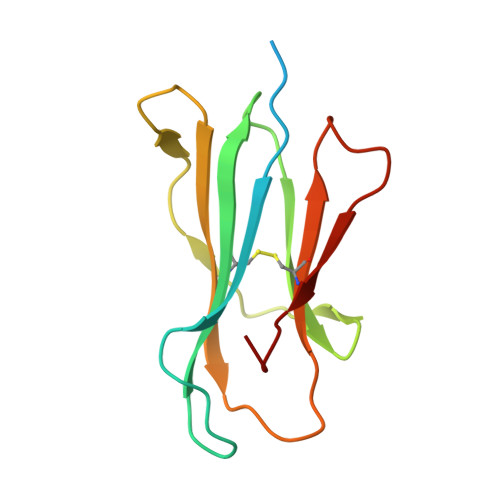
Structural basis for pH-insensitive inhibition of immunoglobulin G recycling by an anti-neonatal Fc receptor antibody.
Kenniston, J.A., Taylor, B.M., Conley, G.P., Cosic, J., Kopacz, K.J., Lindberg, A.P., Comeau, S.R., Atkins, K., Bullen, J., TenHoor, C., Adelman, B.A., Sexton, D.J., Edwards, T.E., Nixon, A.E.(2017) J Biol Chem 292: 17449-17460
- PubMed: 28878017
- DOI: https://doi.org/10.1074/jbc.M117.807396
- Primary Citation of Related Structures:
5WHJ, 5WHK - PubMed Abstract:
The neonatal Fc receptor FcRn plays a critical role in the trafficking of IgGs across tissue barriers and in retaining high circulating concentrations of both IgG and albumin. Although generally beneficial from an immunological perspective in maintaining IgG populations, FcRn can contribute to the pathogenesis of autoimmune disorders when an abnormal immune response targets normal biological components. We previously described a monoclonal antibody (DX-2507) that binds to FcRn with high affinity at both neutral and acidic pH, prevents the simultaneous binding of IgG, and reduces circulating IgG levels in preclinical animal models. Here, we report a 2.5 Å resolution X-ray crystal structure of an FcRn-DX-2507 Fab complex, revealing a nearly complete overlap of the IgG-Fc binding site in FcRn by complementarity-determining regions in DX-2507. This overlap explains how DX-2507 blocks IgG binding to FcRn and thereby shortens IgG half-life by preventing IgGs from recycling back into circulation. Moreover, the complex structure explains how the DX-2507 interaction is pH-insensitive unlike normal Fc interactions and how serum albumin levels are unaffected by DX-2507 binding. These structural studies could inform antibody-based therapeutic approaches for limiting the effects of IgG-mediated autoimmune disease.
Organizational Affiliation:
From Shire, Lexington, Massachusetts 02421, jkenniston0@shire.com.