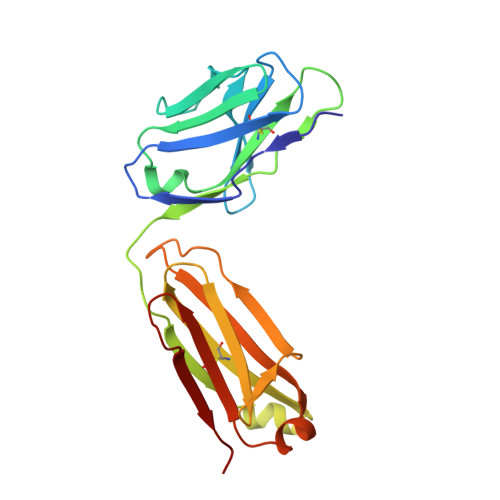
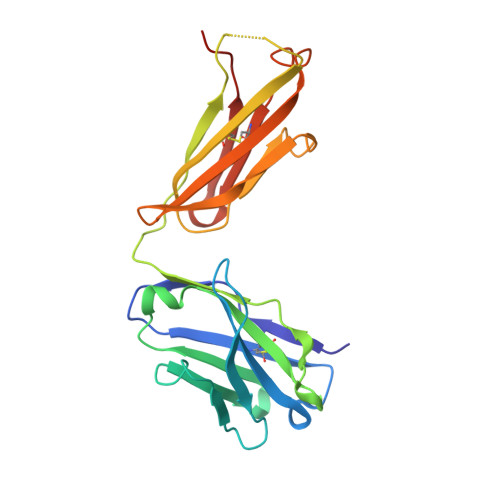
T-dependent B cell responses to Plasmodium induce antibodies that form a high-avidity multivalent complex with the circumsporozoite protein.
Fisher, C.R., Sutton, H.J., Kaczmarski, J.A., McNamara, H.A., Clifton, B., Mitchell, J., Cai, Y., Dups, J.N., D'Arcy, N.J., Singh, M., Chuah, A., Peat, T.S., Jackson, C.J., Cockburn, I.A.(2017) PLoS Pathog 13: e1006469-e1006469
- PubMed: 28759640
- DOI: https://doi.org/10.1371/journal.ppat.1006469
- Primary Citation of Related Structures:
5SZF - PubMed Abstract:
The repeat region of the Plasmodium falciparum circumsporozoite protein (CSP) is a major vaccine antigen because it can be targeted by parasite neutralizing antibodies; however, little is known about this interaction. We used isothermal titration calorimetry, X-ray crystallography and mutagenesis-validated modeling to analyze the binding of a murine neutralizing antibody to Plasmodium falciparum CSP. Strikingly, we found that the repeat region of CSP is bound by multiple antibodies. This repeating pattern allows multiple weak interactions of single FAB domains to accumulate and yield a complex with a dissociation constant in the low nM range. Because the CSP protein can potentially cross-link multiple B cell receptors (BCRs) we hypothesized that the B cell response might be T cell independent. However, while there was a modest response in mice deficient in T cell help, the bulk of the response was T cell dependent. By sequencing the BCRs of CSP-repeat specific B cells in inbred mice we found that these cells underwent somatic hypermutation and affinity maturation indicative of a T-dependent response. Last, we found that the BCR repertoire of responding B cells was limited suggesting that the structural simplicity of the repeat may limit the breadth of the immune response.
Organizational Affiliation:
Research School of Chemistry, The Australian National University, Canberra, Australian Capital Territory, Australia.