Crystal structure of human carbonic anhydrase isozyme XIII with 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide
Smirnov, A., Manakova, E., Grazulis, S., Matulis, D.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Carbonic anhydrase 13 | A [auth B], B [auth A] | 263 | Homo sapiens | Mutation(s): 0 Gene Names: CA13 EC: 4.2.1.1 | 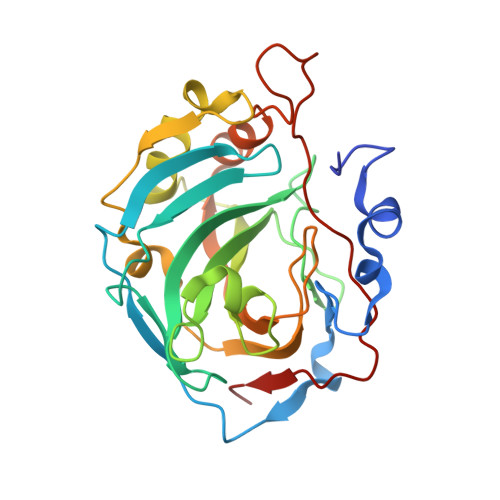 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8N1Q1 (Homo sapiens) Explore Q8N1Q1 Go to UniProtKB: Q8N1Q1 | |||||
PHAROS: Q8N1Q1 GTEx: ENSG00000185015 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8N1Q1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 9 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 5DU Query on 5DU | O [auth B] | 2-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide C17 H17 F3 N2 O3 S2 HYCVHVOZJGSFTR-NSHDSACASA-N |  | ||
| CIT Query on CIT | D [auth B], N [auth B], U [auth A], W [auth A] | CITRIC ACID C6 H8 O7 KRKNYBCHXYNGOX-UHFFFAOYSA-N |  | ||
| BCN Query on BCN | H [auth B] | BICINE C6 H13 N O4 FSVCELGFZIQNCK-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | K [auth B], Q [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| LCP Query on LCP | L [auth B] | PERCHLORATE ION Cl O4 VLTRZXGMWDSKGL-UHFFFAOYSA-M |  | ||
| ZN Query on ZN | C [auth B], P [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | E [auth B] F [auth B] I [auth B] J [auth B] R [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | M [auth B], V [auth A] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| AZI Query on AZI | G [auth B] | AZIDE ION N3 IVRMZWNICZWHMI-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 56.501 | α = 90 |
| b = 57.419 | β = 90 |
| c = 159.909 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| SCALA | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| MOLREP | phasing |