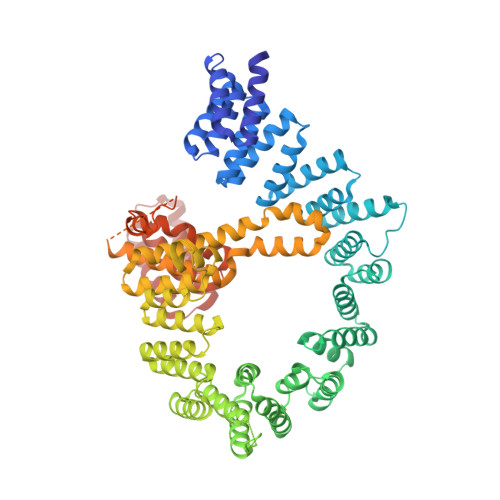
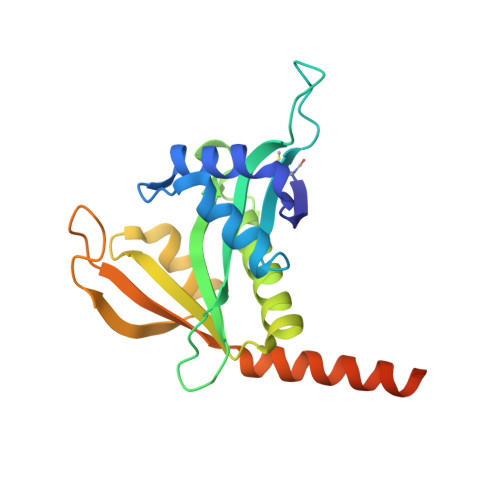
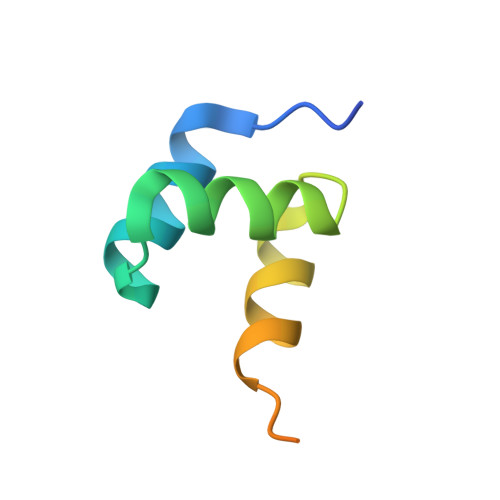
Structural basis of HypK regulating N-terminal acetylation by the NatA complex.
Weyer, F.A., Gumiero, A., Lapouge, K., Bange, G., Kopp, J., Sinning, I.(2017) Nat Commun 8: 15726-15726
- PubMed: 28585574
- DOI: https://doi.org/10.1038/ncomms15726
- Primary Citation of Related Structures:
5NNP, 5NNR - PubMed Abstract:
In eukaryotes, N-terminal acetylation is one of the most common protein modifications involved in a wide range of biological processes. Most N-acetyltransferase complexes (NATs) act co-translationally, with the heterodimeric NatA complex modifying the majority of substrate proteins. Here we show that the Huntingtin yeast two-hybrid protein K (HypK) binds tightly to the NatA complex comprising the auxiliary subunit Naa15 and the catalytic subunit Naa10. The crystal structures of NatA bound to HypK or to a N-terminal deletion variant of HypK were determined without or with a bi-substrate analogue, respectively. The HypK C-terminal region is responsible for high-affinity interaction with the C-terminal part of Naa15. In combination with acetylation assays, the HypK N-terminal region is identified as a negative regulator of the NatA acetylation activity. Our study provides mechanistic insights into the regulation of this pivotal protein modification.
Organizational Affiliation:
Heidelberg University Biochemistry Center (BZH), INF328, D-69120 Heidelberg, Germany.