Computational Design of Epitope-Specific Functional Antibodies.
Nimrod, G., Fischman, S., Austin, M., Herman, A., Keyes, F., Leiderman, O., Hargreaves, D., Strajbl, M., Breed, J., Klompus, S., Minton, K., Spooner, J., Buchanan, A., Vaughan, T.J., Ofran, Y.(2018) Cell Rep 25: 2121-2131.e5
- PubMed: 30463010
- DOI: https://doi.org/10.1016/j.celrep.2018.10.081
- Primary Citation of Related Structures:
5N7W - PubMed Abstract:
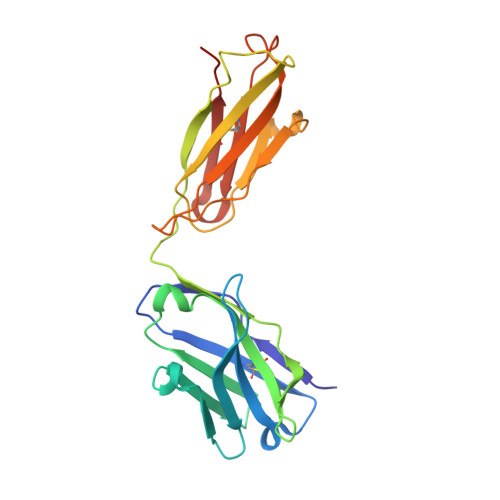
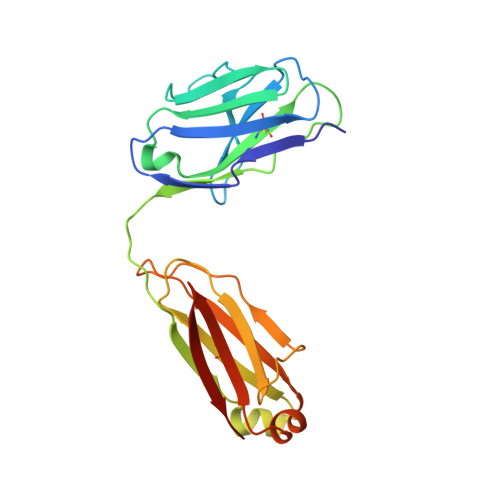
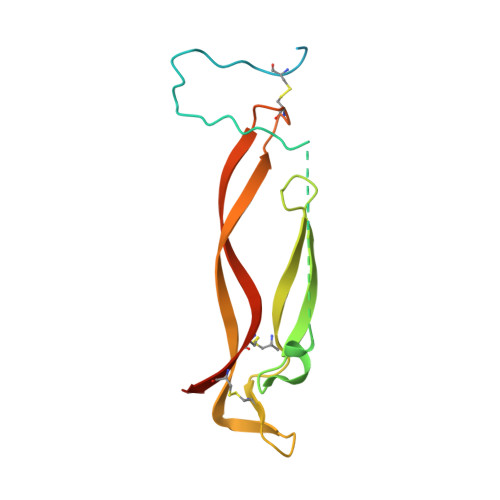
The ultimate goal of protein design is to introduce new biological activity. We propose a computational approach for designing functional antibodies by focusing on functional epitopes, integrating large-scale statistical analysis with multiple structural models. Machine learning is used to analyze these models and predict specific residue-residue contacts. We use this approach to design a functional antibody to counter the proinflammatory effect of the cytokine interleukin-17A (IL-17A). X-ray crystallography confirms that the designed antibody binds the targeted epitope and the interaction is mediated by the designed contacts. Cell-based assays confirm that the antibody is functional. Importantly, this approach does not rely on a high-quality 3D model of the designed complex or even a solved structure of the target. As demonstrated here, this approach can be used to design biologically active antibodies, removing some of the main hurdles in antibody design and in drug discovery.
Organizational Affiliation:
Biolojic Design, Ltd., 12 Hamada Street, Rehovot 7670314, Israel.