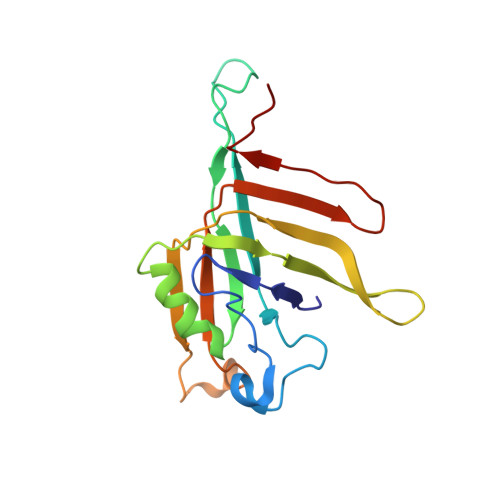
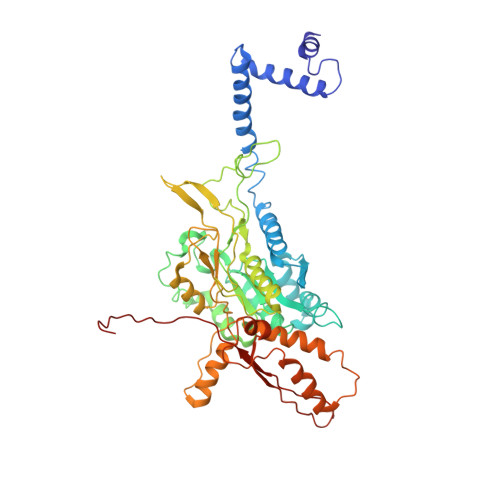
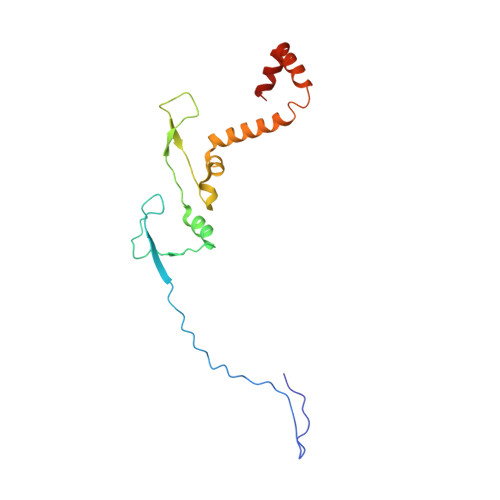
Cryo-EM structure of the extended type VI secretion system sheath-tube complex.
Wang, J., Brackmann, M., Castano-Diez, D., Kudryashev, M., Goldie, K.N., Maier, T., Stahlberg, H., Basler, M.(2017) Nat Microbiol 2: 1507-1512
- PubMed: 28947741
- DOI: https://doi.org/10.1038/s41564-017-0020-7
- Primary Citation of Related Structures:
5MXN, 5MYU, 5OJQ - PubMed Abstract:
The bacterial type VI secretion system (T6SS) uses contraction of a long sheath to quickly thrust a tube with associated effectors across membranes of eukaryotic and bacterial cells 1-5 . Only limited structural information is available about the inherently unstable precontraction state of the T6SS. Here, we obtain a 3.7 Å resolution structure of a non-contractile sheath-tube complex using cryo-electron microscopy and show that it resembles the extended T6SS inside Vibrio cholerae cells. We build a pseudo-atomic model of the complete sheath-tube assembly, which provides a mechanistic understanding of coupling sheath contraction with pushing and rotating the inner tube for efficient target membrane penetration. Our data further show that sheath contraction exposes a buried recognition domain to specifically trigger the disassembly and recycling of the T6SS sheath by the cognate ATP-dependent unfoldase ClpV.
Organizational Affiliation:
Focal Area Infection Biology, Biozentrum, University of Basel, Klingelbergstrasse 50/70, CH-4056, Basel, Switzerland.