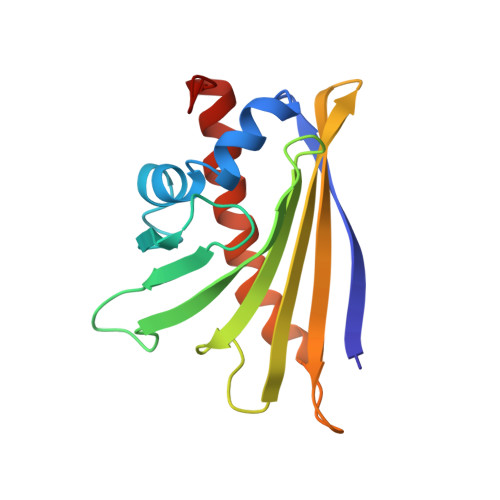
Structure of the Major Apple Allergen Mal d 1.
Ahammer, L., Grutsch, S., Kamenik, A.S., Liedl, K.R., Tollinger, M.(2017) J Agric Food Chem 65: 1606-1612
- PubMed: 28161953
- DOI: https://doi.org/10.1021/acs.jafc.6b05752
- Primary Citation of Related Structures:
5MMU - PubMed Abstract:
More than 70% of birch pollen-allergic patients develop allergic cross-reactions to the major allergen found in apple fruits (Malus domestica), the 17.5 kDa protein Mal d 1. Allergic reactions against this protein result from initial sensitization to the major allergen from birch pollen, Bet v 1. Immunologic cross-reactivity of Bet v 1-specific IgE antibodies with Mal d 1 after apple consumption can subsequently provoke severe oral allergic syndromes. This study presents the three-dimensional NMR solution structure of Mal d 1 (isoform Mal d 1.0101, initially cloned from 'Granny Smith' apples). This protein is composed of a seven-stranded antiparallel β-sheet and three α-helices that form a large internal cavity, similar to Bet v 1 and other cross-reactive food allergens. The Mal d 1 structure provides the basis for elucidating the details of allergic cross-reactivity between birch pollen and apple allergens on a molecular level.
Organizational Affiliation:
Institute of Organic Chemistry, Center for Molecular Biosciences Innsbruck (CMBI), University of Innsbruck , Innrain 80/82, A-6020 Innsbruck, Austria.