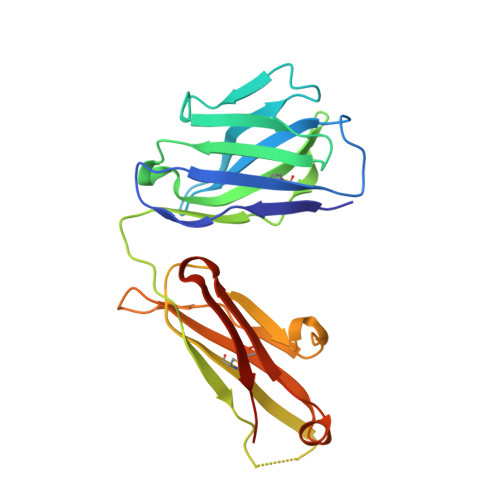
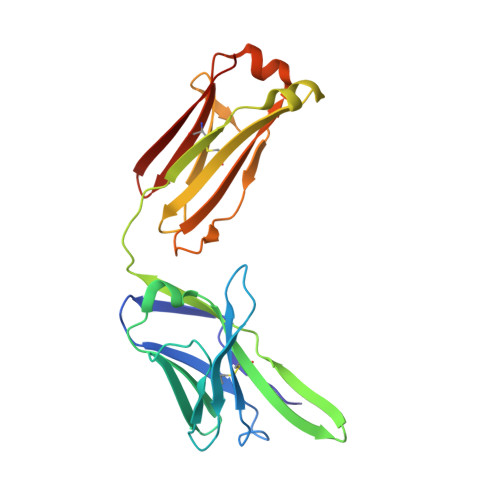
Immunodominance of Antibody Recognition of the HIV Envelope V2 Region in Ig-Humanized Mice.
Wiehe, K., Nicely, N.I., Lockwood, B., Kuraoka, M., Anasti, K., Arora, S., Bowman, C.M., Stolarchuk, C., Parks, R., Lloyd, K.E., Xia, S.M., Duffy, R., Shen, X., Kyratsous, C.A., Macdonald, L.E., Murphy, A.J., Scearce, R.M., Moody, M.A., Alam, S.M., Verkoczy, L., Tomaras, G.D., Kelsoe, G., Haynes, B.F.(2017) J Immunol 198: 1047-1055
- PubMed: 28011932
- DOI: https://doi.org/10.4049/jimmunol.1601640
- Primary Citation of Related Structures:
5KG9 - PubMed Abstract:
In the RV144 gp120 HIV vaccine trial, decreased transmission risk was correlated with Abs that reacted with a linear epitope at a lysine residue at position 169 (K169) in the HIV-1 envelope (Env) V2 region. The K169 V2 response was restricted to Abs bearing Vλ rearrangements that expressed aspartic acid/glutamic acid in CDR L2. The AE.A244 gp120 in AIDSVAX B/E also bound to the unmutated ancestor of a V2-glycan broadly neutralizing Ab, but this Ab type was not induced in the RV144 trial. In this study, we sought to determine whether immunodominance of the V2 linear epitope could be overcome in the absence of human Vλ rearrangements. We immunized IgH- and Igκ-humanized mice with the AE.A244 gp120 Env. In these mice, the V2 Ab response was focused on a linear epitope that did not include K169. V2 Abs were isolated that used the same human VH gene segment as an RV144 V2 Ab but paired with a mouse λ L chain. Structural characterization of one of these V2 Abs revealed how the linear V2 epitope could be engaged, despite the lack of aspartic acid/glutamic acid encoded in the mouse repertoire. Thus, despite the absence of the human Vλ locus in these humanized mice, the dominance of Vλ pairing with human VH for HIV-1 Env V2 recognition resulted in human VH pairing with mouse λ L chains instead of allowing otherwise subdominant V2-glycan broadly neutralizing Abs to develop.
Organizational Affiliation:
Duke Human Vaccine Institute, Duke University School of Medicine, Durham, NC 27710; kevin.wiehe@dm.duke.edu barton.haynes@dm.duke.edu.