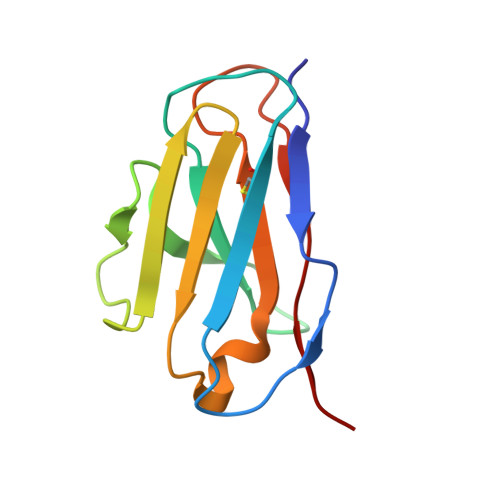
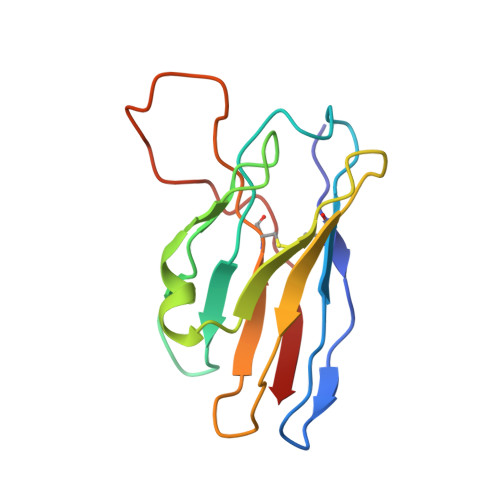
Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Pallesen, J., Murin, C.D., de Val, N., Cottrell, C.A., Hastie, K.M., Turner, H.L., Fusco, M.L., Flyak, A.I., Zeitlin, L., Crowe, J.E., Andersen, K.G., Saphire, E.O., Ward, A.B.(2016) Nat Microbiol 1: 16128-16128
- PubMed: 27562261
- DOI: https://doi.org/10.1038/nmicrobiol.2016.128
- Primary Citation of Related Structures:
5KEM - PubMed Abstract:
The Ebola virus (EBOV) GP gene encodes two glycoproteins. The major product is a soluble, dimeric glycoprotein (sGP) that is secreted abundantly. Despite the abundance of sGP during infection, little is known regarding its structure or functional role. A minor product, resulting from transcriptional editing, is the transmembrane-anchored, trimeric viral surface glycoprotein (GP). GP mediates attachment to and entry into host cells, and is the intended target of antibody therapeutics. Because large portions of sequence are shared between GP and sGP, it has been hypothesized that sGP may potentially subvert the immune response or may contribute to pathogenicity. In this study, we present cryo-electron microscopy structures of GP and sGP in complex with GP-specific and GP/sGP cross-reactive antibodies undergoing human clinical trials. The structure of the sGP dimer presented here, in complex with both an sGP-specific antibody and a GP/sGP cross-reactive antibody, permits us to unambiguously assign the oligomeric arrangement of sGP and compare its structure and epitope presentation to those of GP. We also provide biophysical evaluation of naturally occurring GP/sGP mutations that fall within the footprints identified by our high-resolution structures. Taken together, our data provide a detailed and more complete picture of the accessible Ebolavirus glycoprotein landscape and a structural basis to evaluate patient and vaccine antibody responses towards differently structured products of the GP gene.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, California 92037, USA.