5HBV
Complex structure of Fab35 and mouse nAChR alpha1
- PDB DOI: https://doi.org/10.2210/pdb5HBV/pdb
- Classification: TRANSPORT PROTEIN/TOXIN
- Organism(s): Bungarus multicinctus, Mus musculus, Rattus norvegicus
- Expression System: Pichia
- Mutation(s): Yes
- Deposited: 2016-01-02 Released: 2017-05-03
Experimental Data Snapshot
- Method: X-RAY DIFFRACTION
- Resolution: 2.70 Å
- R-Value Free: 0.268
- R-Value Work: 0.227
- R-Value Observed: 0.229
wwPDB Validation 3D Report Full Report
This is version 2.0 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Alpha-bungarotoxin isoform V31 | 74 | Bungarus multicinctus | Mutation(s): 0 | 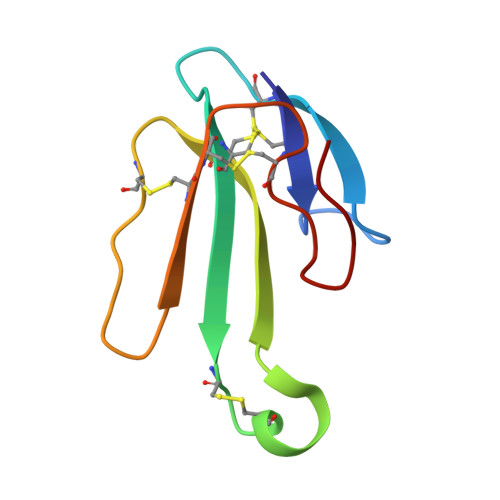 | |
UniProt | |||||
Find proteins for P60616 (Bungarus multicinctus) Explore P60616 Go to UniProtKB: P60616 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60616 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetylcholine receptor subunit alpha 1 | 212 | Mus musculus | Mutation(s): 3 Gene Names: Chrna1, Acra | 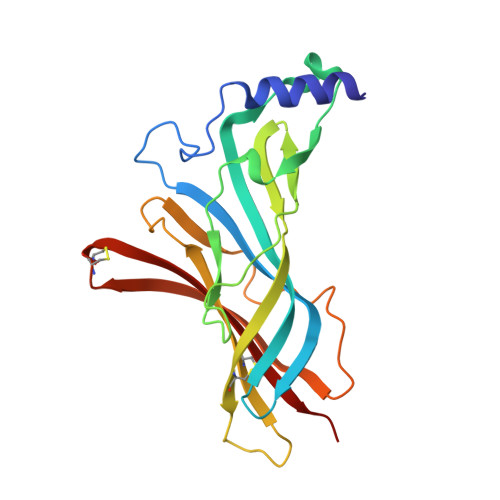 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P04756 (Mus musculus) Explore P04756 Go to UniProtKB: P04756 | |||||
IMPC: MGI:87885 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04756 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fab35, Light Chain | 213 | Rattus norvegicus | Mutation(s): 0 | 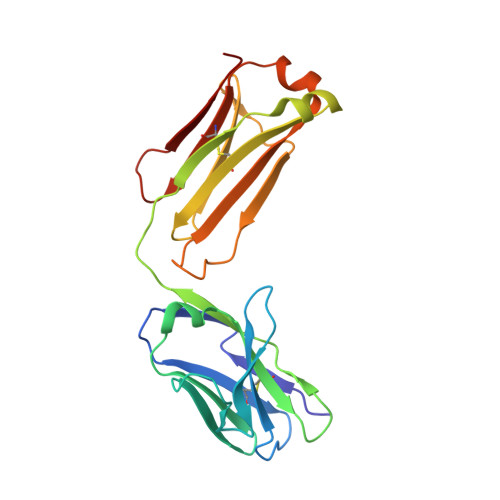 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fab35, Heavy Chain | 219 | Rattus norvegicus | Mutation(s): 0 | 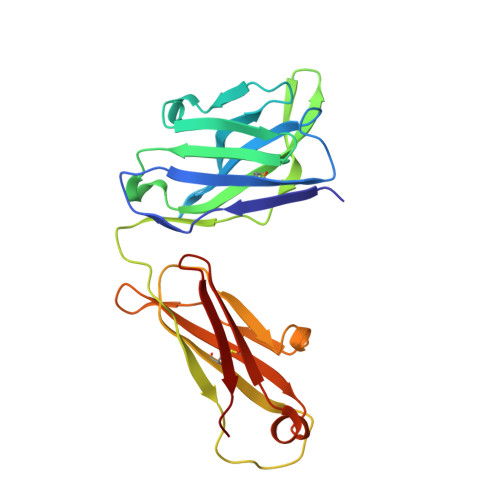 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Oligosaccharides
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | E | 9 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G68668TB GlyCosmos: G68668TB GlyGen: G68668TB | |||||
Experimental Data & Validation
Experimental Data
- Method: X-RAY DIFFRACTION
- Resolution: 2.70 Å
- R-Value Free: 0.268
- R-Value Work: 0.227
- R-Value Observed: 0.229
- Space Group: C 1 2 1
Unit Cell:
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 159.905 | α = 90 |
| b = 42.024 | β = 116.46 |
| c = 137.583 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data scaling |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHASER | phasing |
Entry History
Deposition Data
- Released Date: 2017-05-03 Deposition Author(s): Noridomi, K., Watanabe, G., Hansen, M.N., Han, G.W., Chen, L.
Revision History (Full details and data files)
- Version 1.0: 2017-05-03
Type: Initial release - Version 1.1: 2017-05-10
Changes: Database references - Version 2.0: 2020-07-29
Type: Remediation
Reason: Carbohydrate remediation
Changes: Atomic model, Data collection, Derived calculations, Structure summary













