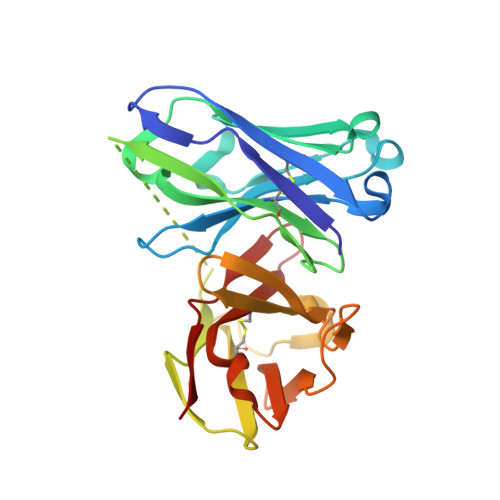
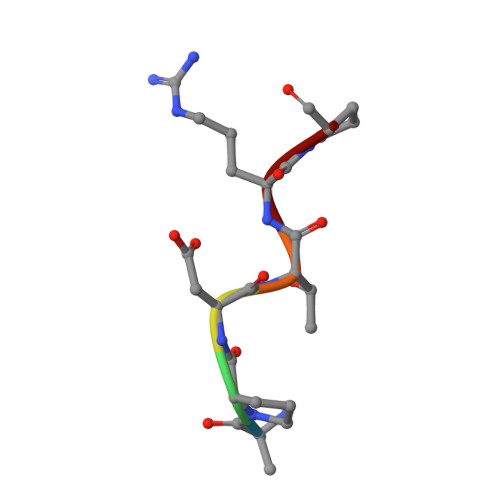
Design of Alpha-S-Glycopeptides Derived from Muc1 with a Flexible and Solvent Exposed Sugar Moiety
Rojas-Ocariz, V., Companon, I., Aydillo, C., Castro-Lopez, J., Jimenez-Barbero, J., Hurtado-Guerrero, R., Avenoza, A., Zurbano, M.M., Corzana, F., Busto, J.H., Peregrina, J.M.(2016) J Org Chem 81: 5929
- PubMed: 27305427
- DOI: https://doi.org/10.1021/acs.joc.6b00833
- Primary Citation of Related Structures:
5FXC - PubMed Abstract:
The use of vaccines based on MUC1 glycopeptides is a promising approach to treat cancer. We present herein several sulfa-Tn antigens incorporated in MUC1 sequences that possess a variable linker between the carbohydrate (GalNAc) and the peptide backbone. The main conformations of these molecules in solution have been evaluated by combining NMR experiments and molecular dynamics simulations. The linker plays a key role in the modulation of the conformation of these compounds at different levels, blocking a direct contact between the sugar moiety and the backbone, promoting a helix-like conformation for the glycosylated residue and favoring the proper presentation of the sugar unit for molecular recognition events. The feasibility of these novel compounds as mimics of MUC1 antigens has been validated by the X-ray diffraction structure of one of these unnatural derivatives complexed to an anti-MUC1 monoclonal antibody. These features, together with potential lack of immune suppression, render these unnatural glycopeptides promising candidates for designing alternative therapeutic vaccines against cancer.
Organizational Affiliation:
Departamento de Química, Centro de Investigación en Síntesis Química, Universidad de La Rioja , Madre de Dios 53, 26006 Logroño, Spain.