Structural investigation and inhibitory response of halide on phosphoserine aminotransferase from Trichomonas vaginalis.
Singh, R.K., Mazumder, M., Sharma, B., Gourinath, S.(2016) Biochim Biophys Acta 1860: 1508-1518
- PubMed: 27102280
- DOI: https://doi.org/10.1016/j.bbagen.2016.04.013
- Primary Citation of Related Structures:
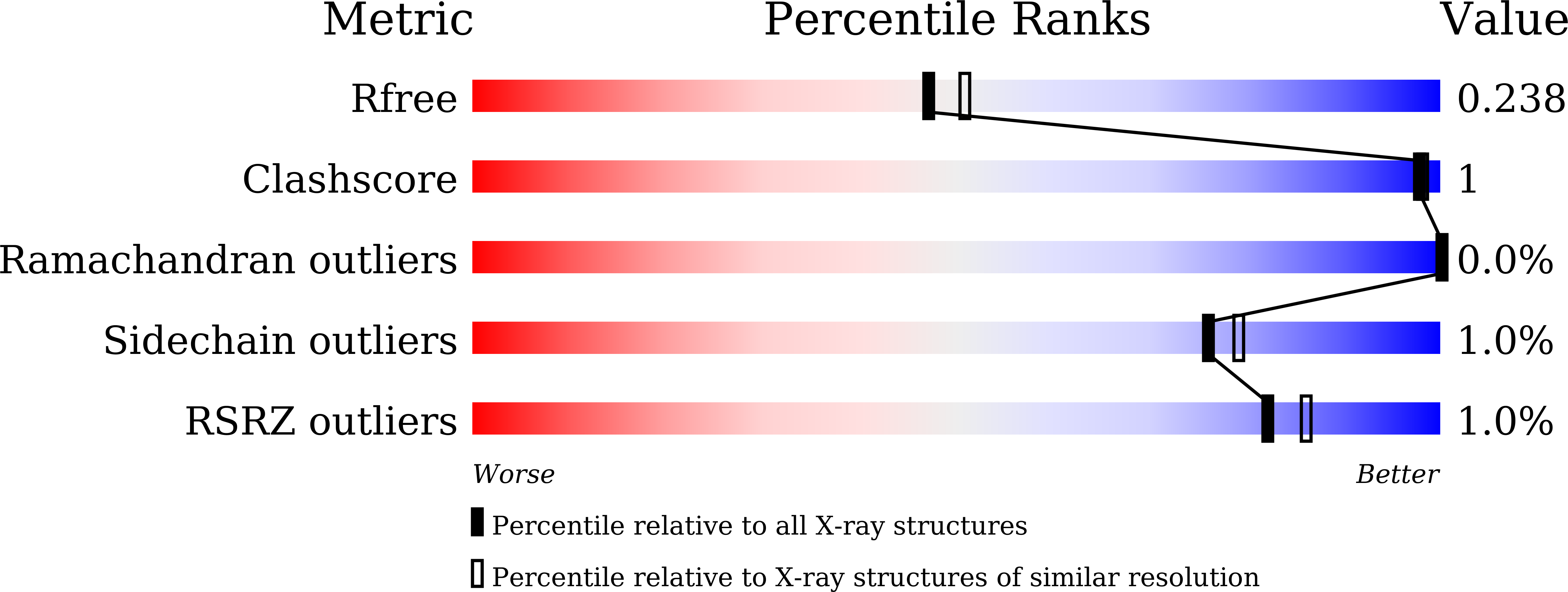
5F8V - PubMed Abstract:
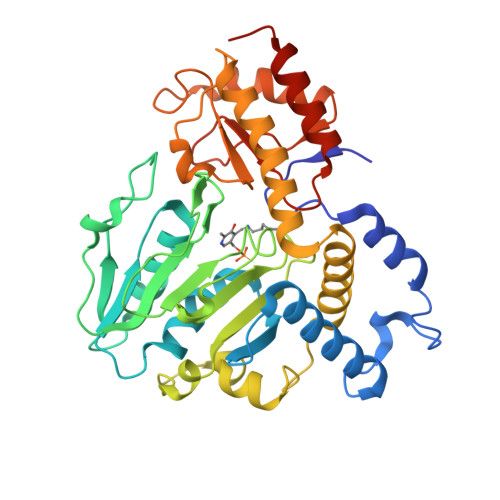
Phosphoserine aminotransferase (PSAT) catalyses the second reversible step of the phosphoserine biosynthetic pathway in Trichomonas vaginalis, which is crucial for the synthesis of serine and cysteine. PSAT from T. vaginalis (TvPSAT) was analysed using X-ray crystallography, enzyme kinetics, and molecular dynamics simulations. The crystal structure of TvPSAT was determined to 2.15Å resolution, and is the first protozoan PSAT structure to be reported. The active site of TvPSAT structure was found to be in a closed conformation, and at the active site PLP formed an internal aldimine linkage to Lys 202. In TvPSAT, Val 340 near the active site while it is Arg in most other members of the PSAT family, might be responsible in closing the active site. Kinetic studies yielded Km values of 54 μM and 202 μM for TvPSAT with OPLS and AKG, respectively. Only iodine inhibited the TvPSAT activity while smaller halides could not inhibit. Results from the structure, comparative molecular dynamics simulations, and the inhibition studies suggest that iodine is the only halide that can bind TvPSAT strongly and may thus inhibit the activity of TvPSAT. The long loop between β8 and α8 at the opening of the TvPSAT active site cleft compared to other PSATs, suggests that this loop may help control the access of substrates to the TvPSAT active site and thus influences the enzyme kinetics. Our structural and functional studies have improved our understanding of how PSAT helps this organism persists in the environment.
Organizational Affiliation:
School of Life Sciences, Jawaharlal Nehru University, New Delhi, India.