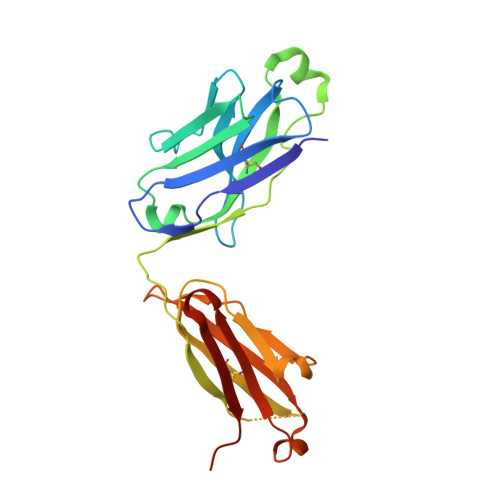
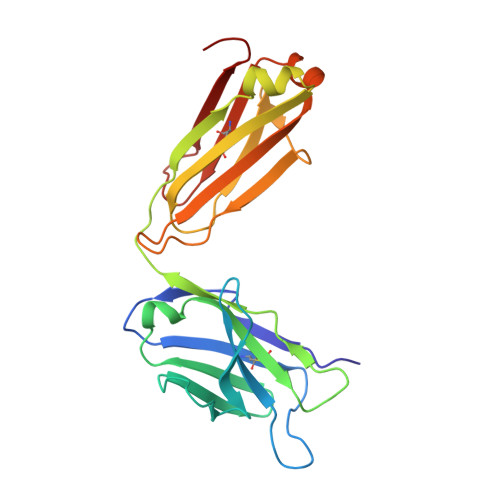
Distinct homotypic B-cell receptor interactions shape the outcome of chronic lymphocytic leukaemia.
Minici, C., Gounari, M., Ubelhart, R., Scarfo, L., Duhren-von Minden, M., Schneider, D., Tasdogan, A., Alkhatib, A., Agathangelidis, A., Ntoufa, S., Chiorazzi, N., Jumaa, H., Stamatopoulos, K., Ghia, P., Degano, M.(2017) Nat Commun 8: 15746-15746
- PubMed: 28598442
- DOI: https://doi.org/10.1038/ncomms15746
- Primary Citation of Related Structures:
5DRW, 5DRX, 5IFH - PubMed Abstract:
Cell-autonomous B-cell receptor (BcR)-mediated signalling is a hallmark feature of the neoplastic B lymphocytes in chronic lymphocytic leukaemia (CLL). Here we elucidate the structural basis of autonomous activation of CLL B cells, showing that BcR immunoglobulins initiate intracellular signalling through homotypic interactions between epitopes that are specific for each subgroup of patients with homogeneous clinicobiological profiles. The molecular details of the BcR-BcR interactions apparently dictate the clinical course of disease, with stronger affinities and longer half-lives in indolent cases, and weaker, short-lived contacts mediating the aggressive ones. The diversity of homotypic BcR contacts leading to cell-autonomous signalling reconciles the existence of a shared pathogenic mechanism with the biological and clinical heterogeneity of CLL and offers opportunities for innovative treatment strategies.
Organizational Affiliation:
Biocrystallography Unit, Division of Immunology, Transplantation and Infectious Diseases, IRCCS San Raffaele Scientific Institute, via Olgettina 58, 20132 Milan, Italy.