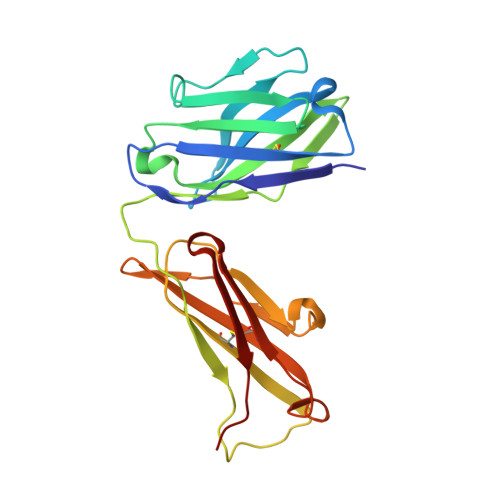
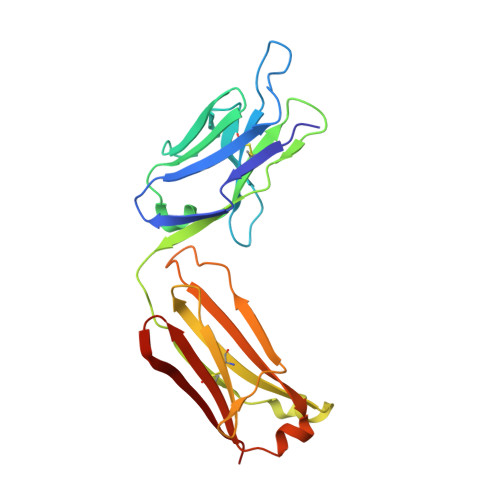
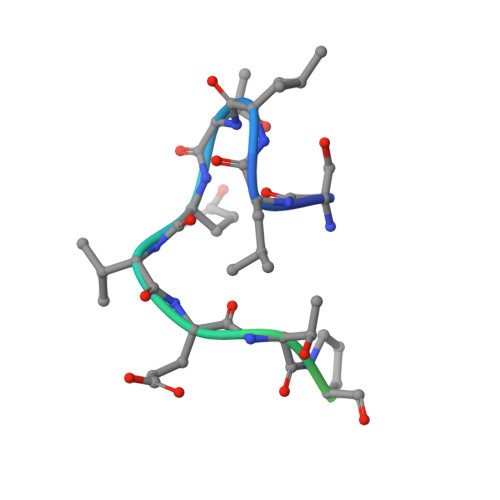
Crystal Structure of the Conserved Amino Terminus of the Extracellular Domain of Matrix Protein 2 of Influenza A Virus Gripped by an Antibody.
Cho, K.J., Schepens, B., Moonens, K., Deng, L., Fiers, W., Remaut, H., Saelens, X.(2015) J Virol 90: 611-615
- PubMed: 26468526
- DOI: https://doi.org/10.1128/JVI.02105-15
- Primary Citation of Related Structures:
5DLM - PubMed Abstract:
We report the crystal structure of the M2 ectodomain (M2e) in complex with a monoclonal antibody that binds the amino terminus of M2. M2e extends into the antibody binding site to form an N-terminal β-turn near the bottom of the paratope. This M2e folding differs significantly from that of M2e in complex with an antibody that binds another part of M2e. This suggests that M2e can adopt at least two conformations that can elicit protective antibodies.
Organizational Affiliation:
Medical Biotechnology Center, VIB, Ghent, Belgium Department of Biomedical Molecular Biology, Ghent University, Ghent, Belgium.