Crystal structure of insect thioredoxin at 1.95 Angstroms
Klinke, S., Tejedor, M.D., Cerutti, M.L., Giacometti, R., Otero, L.H., Goldbaum, F.A., Zavala, J.A., Wolosiuk, R.A., Pagano, E.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Thioredoxin | 109 | Anticarsia gemmatalis | Mutation(s): 0 | 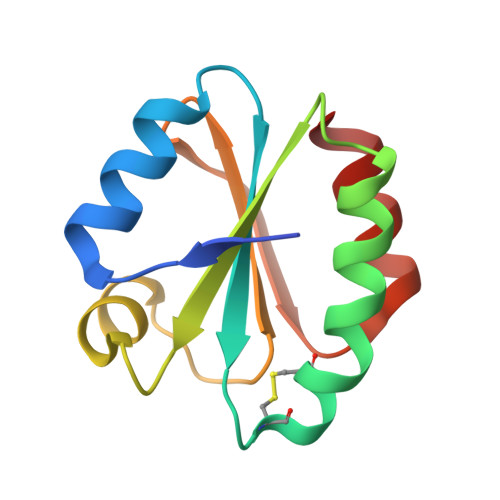 | |
UniProt | |||||
Find proteins for A0A0Y0BMD0 (Anticarsia gemmatalis) Explore A0A0Y0BMD0 Go to UniProtKB: A0A0Y0BMD0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0Y0BMD0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 107.71 | α = 90 |
| b = 28.98 | β = 128.32 |
| c = 79.86 | γ = 90 |
| Software Name | Purpose |
|---|---|
| BUSTER | refinement |
| PROTEUM PLUS | data collection |
| XDS | data scaling |
| MrBUMP | phasing |
| MOLREP | phasing |
| PHASER | phasing |
| Coot | model building |
| XDS | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| ANPCyT | Argentina | PICT 2011-2115 |
| ANPCyT | Argentina | PPL2-03 |