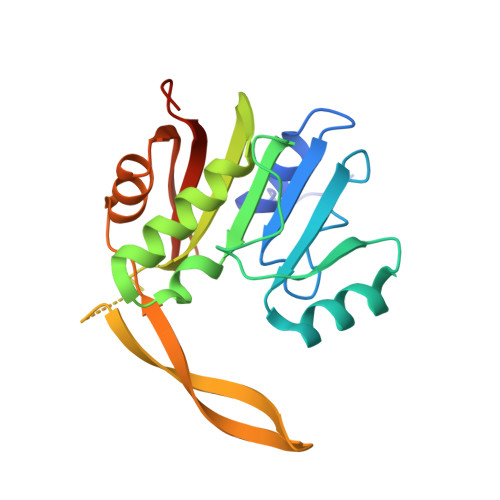
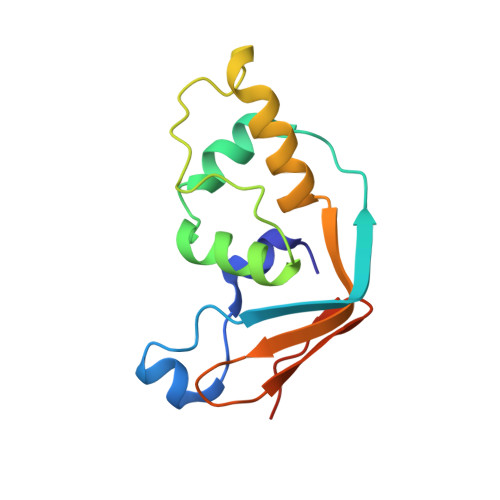
Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure.
Letoquart, J., van Tran, N., Caroline, V., Aleksandrov, A., Lazar, N., van Tilbeurgh, H., Liger, D., Graille, M.(2015) Nucleic Acids Res 43: 10989-11002
- PubMed: 26438534
- DOI: https://doi.org/10.1093/nar/gkv1009
- Primary Citation of Related Structures:
5CM2 - PubMed Abstract:
Most of the factors involved in translation (tRNA, rRNA and proteins) are subject to post-transcriptional and post-translational modifications, which participate in the fine-tuning and tight control of ribosome and protein synthesis processes. In eukaryotes, Trm112 acts as an obligate activating platform for at least four methyltransferases (MTase) involved in the modification of 18S rRNA (Bud23), tRNA (Trm9 and Trm11) and translation termination factor eRF1 (Mtq2). Trm112 is then at a nexus between ribosome synthesis and function. Here, we present a structure-function analysis of the Trm9-Trm112 complex, which is involved in the 5-methoxycarbonylmethyluridine (mcm(5)U) modification of the tRNA anticodon wobble position and hence promotes translational fidelity. We also compare the known crystal structures of various Trm112-MTase complexes, highlighting the structural plasticity allowing Trm112 to interact through a very similar mode with its MTase partners, although those share less than 20% sequence identity.
Organizational Affiliation:
Laboratoire de Biochimie, CNRS, UMR 7654, Ecole Polytechnique, F-91128 Palaiseau Cedex, France Fonction et Architecture des Assemblages Macromoléculaires, Département B3S, Institut de Biologie Intégrative de la Cellule (I2BC), CNRS, UMR 9198, CEA, Université Paris Sud, F-91405 Orsay Cedex, France.