X-ray structure and inhibition of the feline infectious peritonitis virus 3C-like protease: Structural implications for drug design.
St John, S.E., Therkelsen, M.D., Nyalapatla, P.R., Osswald, H.L., Ghosh, A.K., Mesecar, A.D.(2015) Bioorg Med Chem Lett 25: 5072-5077
- PubMed: 26592814
- DOI: https://doi.org/10.1016/j.bmcl.2015.10.023
- Primary Citation of Related Structures:
4ZRO - PubMed Abstract:
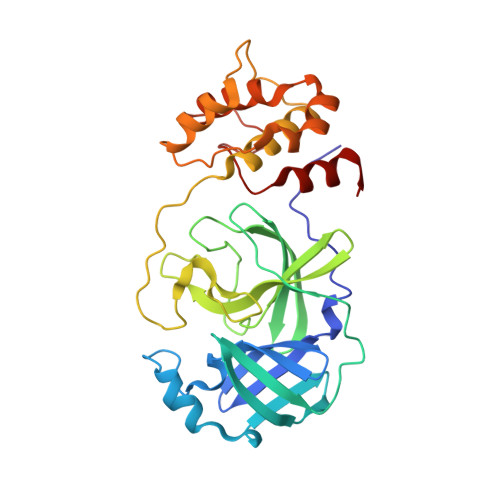
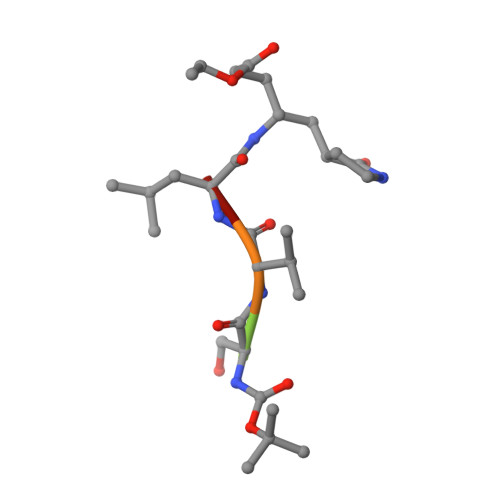
Feline infectious peritonitis (FIP) is a deadly disease that effects both domestic and wild cats and is caused by a mutation in feline coronavirus (FCoV) that allows the virus to replicate in macrophages. Currently, there are no treatments or vaccines available for the treatment of FIP even though it kills approximately 5% of cats in multi-cat households per year. In an effort to develop small molecule drugs targeting FIP for the treatment of cats, we screened a small set of designed peptidomimetic inhibitors for inhibition of FIPV-3CL(pro), identifying two compounds with low to sub-micromolar inhibition, compound 6 (IC50=0.59±0.06 μM) and compound 7 (IC50=1.3±0.1 μM). We determined the first X-ray crystal structure of FIPV-3CL(pro) in complex with the best inhibitor identified, compound 6, to a resolution of 2.10 Å to better understand the structural basis for inhibitor specificity. Our study provides important insights into the structural requirements for the inhibition of FIPV-3CL(pro) by peptidomimetic inhibitors and expands the current structural knowledge of coronaviral 3CL(pro) architecture.
Organizational Affiliation:
Departments of Biochemistry and Biological Sciences, Purdue University, West Lafayette, IN, USA; Centers for Cancer Research & Drug Discovery, Purdue University, West Lafayette, IN, USA.