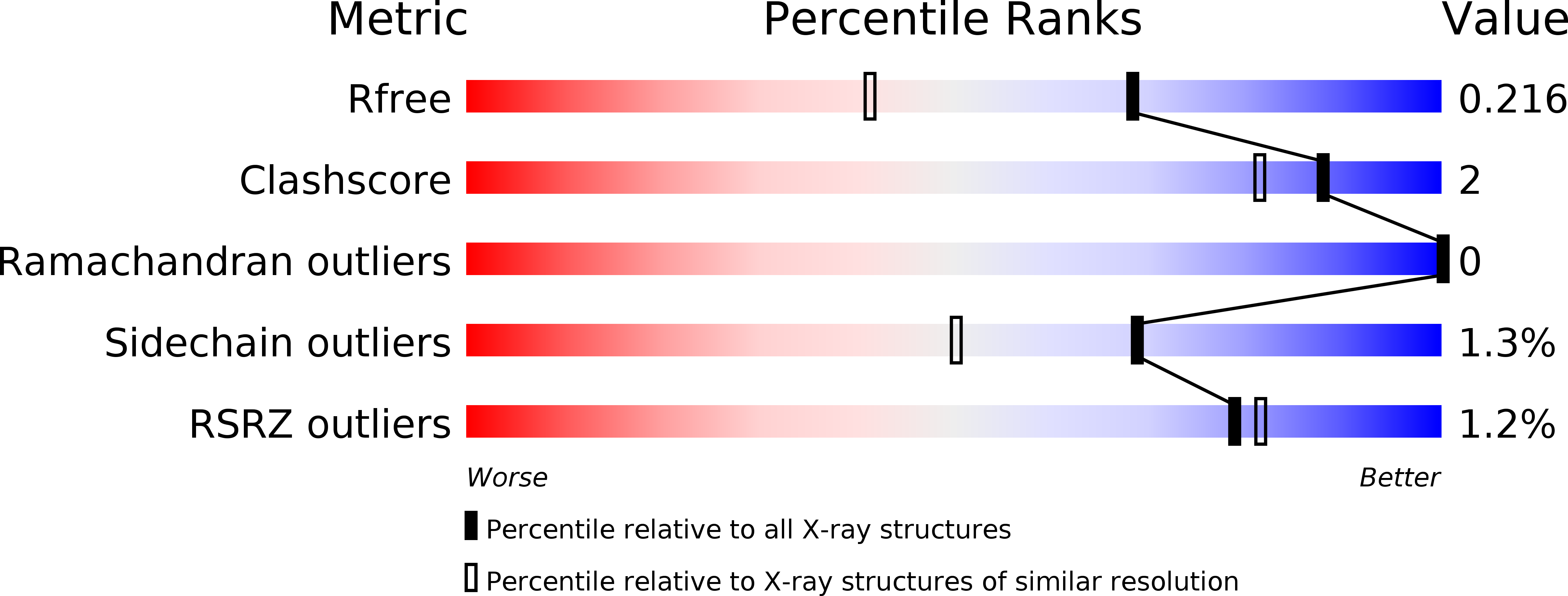
Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex.
Clark, M.D., Marcum, R., Graveline, R., Chan, C.W., Xie, T., Chen, Z., Ding, Y., Zhang, Y., Mondragon, A., David, G., Radhakrishnan, I.(2015) Proc Natl Acad Sci U S A 112: E3669-E3678
- PubMed: 26124119
- DOI: https://doi.org/10.1073/pnas.1504021112
- Primary Citation of Related Structures:
2N2H, 4ZQA - PubMed Abstract:
Acetylation is correlated with chromatin decondensation and transcriptional activation, but its regulation by histone deacetylase (HDAC)-bearing corepressor complexes is poorly understood. Here, we describe the mechanism of assembly of the mammalian Sin3L/Rpd3L complex facilitated by Sds3, a conserved subunit deemed critical for proper assembly. Sds3 engages a globular, helical region of the HDAC interaction domain (HID) of the scaffolding protein Sin3A through a bipartite motif comprising a helix and an adjacent extended segment. Sds3 dimerizes through not only one of the predicted coiled-coil motifs but also, the segment preceding it, forming an ∼ 150-Å-long antiparallel dimer. Contrary to previous findings in yeast, Sin3A rather than Sds3 functions in recruiting HDAC1 into the complex by engaging the latter through a highly conserved segment adjacent to the helical HID subdomain. In the resulting model for the ternary complex, the two copies of the HDACs are situated distally and dynamically because of a natively unstructured linker connecting the dimerization domain and the Sin3A interaction domain of Sds3; these features contrast with the static organization described previously for the NuRD (nucleosome remodeling and deacetylase) complex. The Sds3 linker features several conserved basic residues that could potentially maintain the complex on chromatin by nonspecific interactions with DNA after initial recruitment by sequence-specific DNA-binding repressors.
Organizational Affiliation:
Department of Molecular Biosciences, Northwestern University, Evanston, IL 60208-3500;