Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Rubinstein, R., Thu, C.A., Goodman, K.M., Wolcott, H.N., Bahna, F., Mannepalli, S., Ahlsen, G., Chevee, M., Halim, A., Clausen, H., Maniatis, T., Shapiro, L., Honig, B.(2015) Cell 163: 629-642
- PubMed: 26478182
- DOI: https://doi.org/10.1016/j.cell.2015.09.026
- Primary Citation of Related Structures:
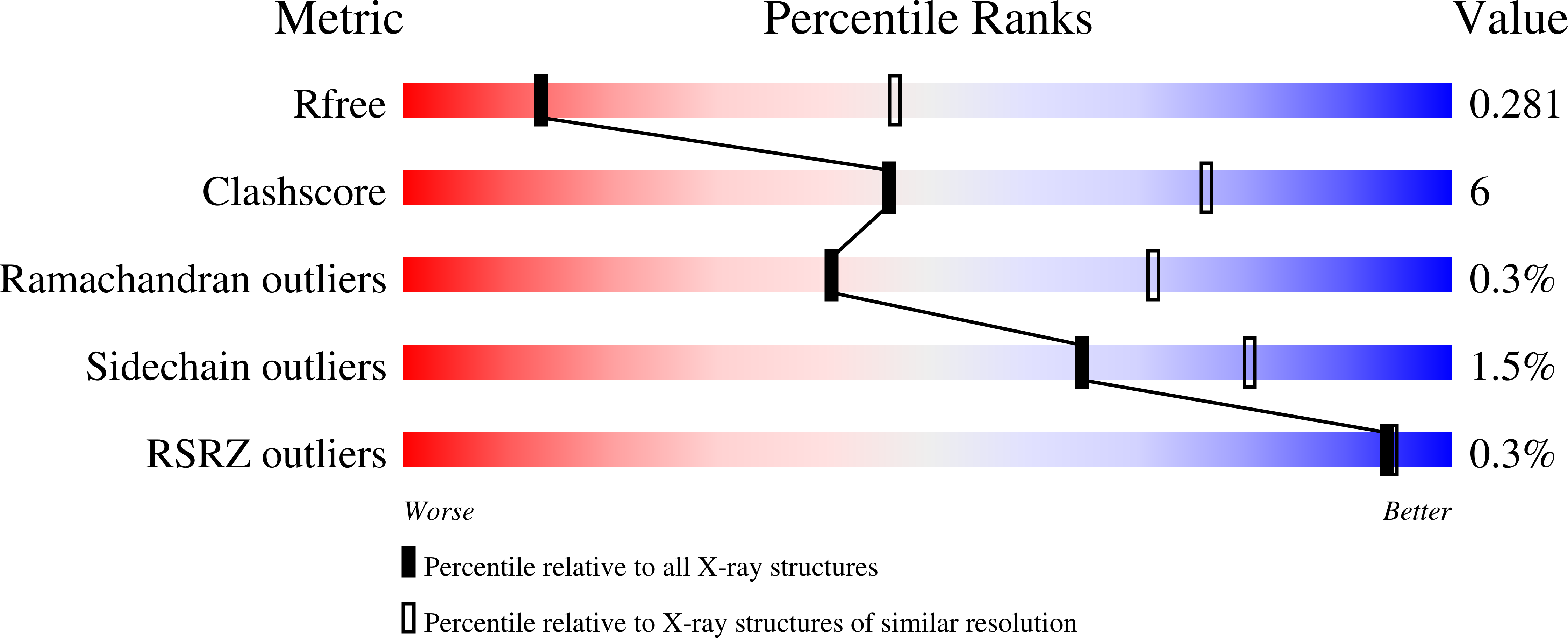
4ZPL, 4ZPM, 4ZPN, 4ZPO, 4ZPP, 4ZPQ, 4ZPS - PubMed Abstract:
Self-avoidance, a process preventing interactions of axons and dendrites from the same neuron during development, is mediated in vertebrates through the stochastic single-neuron expression of clustered protocadherin protein isoforms. Extracellular cadherin (EC) domains mediate isoform-specific homophilic binding between cells, conferring cell recognition through a poorly understood mechanism. Here, we report crystal structures for the EC1-EC3 domain regions from four protocadherin isoforms representing the α, β, and γ subfamilies. All are rod shaped and monomeric in solution. Biophysical measurements, cell aggregation assays, and computational docking reveal that trans binding between cells depends on the EC1-EC4 domains, which interact in an antiparallel orientation. We also show that the EC6 domains are required for the formation of cis-dimers. Overall, our results are consistent with a model in which protocadherin cis-dimers engage in a head-to-tail interaction between EC1-EC4 domains from apposed cell surfaces, possibly forming a zipper-like protein assembly, and thus providing a size-dependent self-recognition mechanism.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA; Department of Systems Biology, Columbia University, New York, NY 10032, USA.