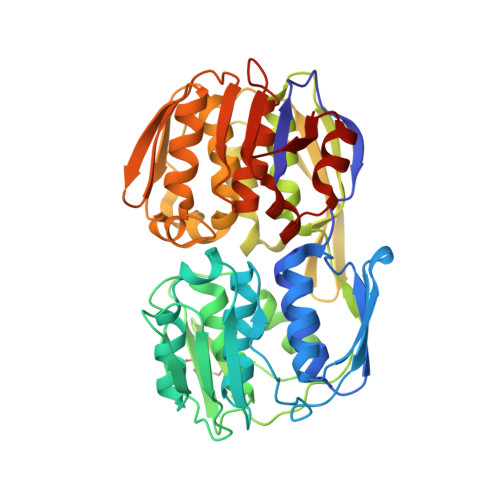
2.55 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate, phosphate, and potassium
Light, S.H., Minasov, G., Krishna, S.N., Kwon, K., Anderson, W.F.To be published.