Crystal structure of the bacteriophage G20C portal protein
Williams, L.S., Turkenburg, J.P., Levdikov, V.M., Minakhin, L., Severinov, K., Antson, A.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Portal protein | 426 | Bacteriophage sp. | Mutation(s): 0 | 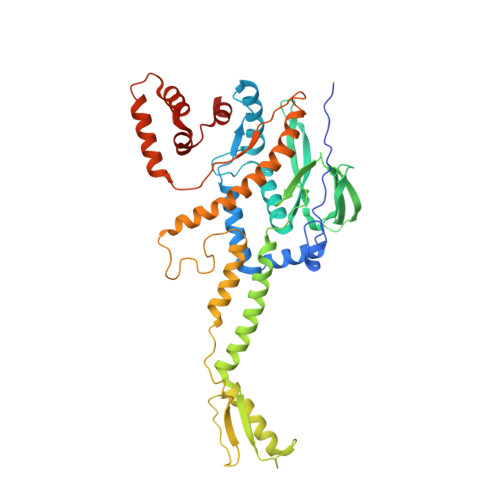 | |
UniProt | |||||
Find proteins for A7XXR3 (Thermus virus P74-26) Explore A7XXR3 Go to UniProtKB: A7XXR3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A7XXR3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MPD Query on MPD | D [auth A], E [auth B], F [auth C] | (4S)-2-METHYL-2,4-PENTANEDIOL C6 H14 O2 SVTBMSDMJJWYQN-YFKPBYRVSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 155.328 | α = 90 |
| b = 155.328 | β = 90 |
| c = 115.445 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Aimless | data scaling |
| XDS | data reduction |
| SHELX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 098230 |