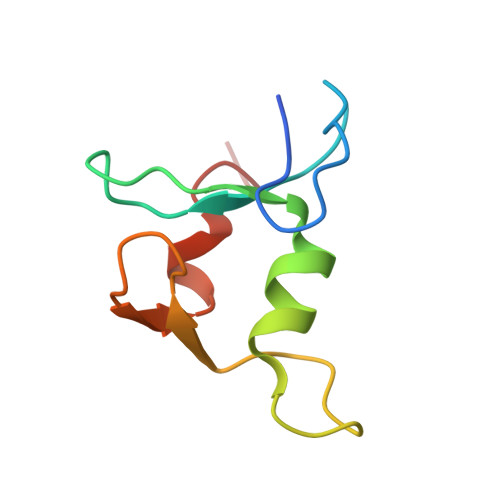
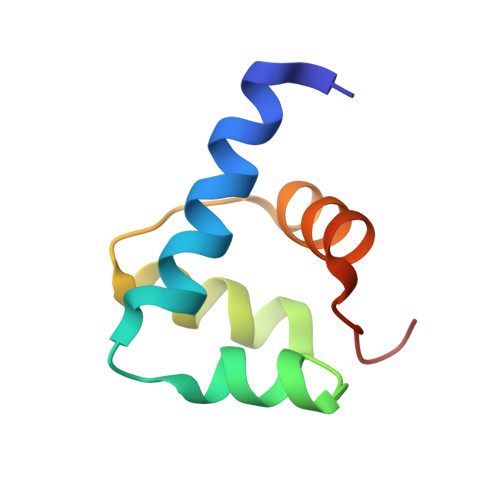
Crystal structure and SUMO binding of Slx1-Slx4 complex
Lian, F.M., Xie, S., Qian, C.M.(2016) Sci Rep 6: 19331-19331
- PubMed: 26787556
- DOI: https://doi.org/10.1038/srep19331
- Primary Citation of Related Structures:
4ZDT - PubMed Abstract:
The SLX1-SLX4 complex is a structure-specific endonuclease that cleaves branched DNA structures and plays significant roles in DNA recombination and repair in eukaryotic cells. The heterodimeric interaction between SLX1 and SLX4 is essential for the endonuclease activity of SLX1. Here, we present the crystal structure of Slx1 C-terminal zinc finger domain in complex with the C-terminal helix-turn-helix domain of Slx4 from Schizosaccharomyces pombe at 2.0 Å resolution. The structure reveals a conserved binding mechanism underling the Slx1-Slx4 interaction. Structural and sequence analyses indicate Slx1 C-terminal domain is actually an atypical C4HC3-type RING finger which normally possesses E3 ubiquitin ligase activity, but here is absolutely required for Slx1 interaction with Slx4. Furthermore, we found the C-terminal tail of S. pombe Slx1 contains a SUMO-interacting motif and can recognize Pmt3 (S. pombe SUMO), suggesting that Slx1-Slx4 complex could be recruited by SUMOylated protein targets to take part in replication associated DNA repair processes.
Organizational Affiliation:
School of Biomedical Sciences, The University of Hong Kong, Hong Kong, China.