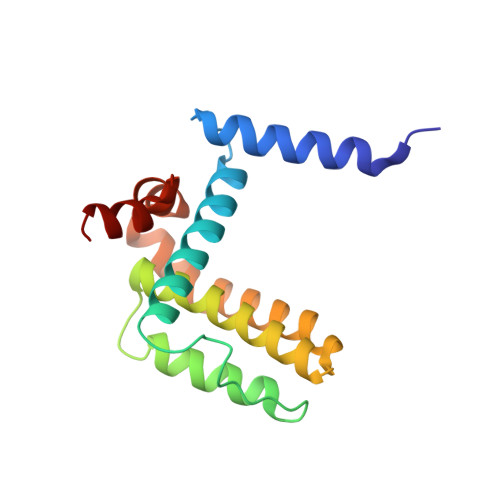
TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL.
Thebault, S., Agez, M., Chi, X., Stojko, J., Cura, V., Telerman, S.B., Maillet, L., Gautier, F., Billas-Massobrio, I., Birck, C., Troffer-Charlier, N., Karafin, T., Honore, J., Senff-Ribeiro, A., Montessuit, S., Johnson, C.M., Juin, P., Cianferani, S., Martinou, J.C., Andrews, D.W., Amson, R., Telerman, A., Cavarelli, J.(2016) Sci Rep 6: 19725-19725
- PubMed: 26813996
- DOI: https://doi.org/10.1038/srep19725
- Primary Citation of Related Structures:
4Z9V - PubMed Abstract:
Translationally Controlled Tumor Protein (TCTP) is anti-apoptotic, key in development and cancer, however without the typical Bcl2 family members' structure. Here we report that TCTP contains a BH3-like domain and forms heterocomplexes with Bcl-xL. The crystal structure of a Bcl-xL deletion variant-TCTP11-31 complex reveals that TCTP refolds in a helical conformation upon binding the BH3-groove of Bcl-xL, although lacking the h1-subregion interaction. Experiments using in vitro-vivo reconstituted systems and TCTP(+/-) mice indicate that TCTP activates the anti-apoptotic function of Bcl-xL, in contrast to all other BH3-proteins. Replacing the non-conserved h1 of TCTP by that of Bax drastically increases the affinity of this hybrid for Bcl-xL, modifying its biological properties. This work reveals a novel class of BH3-proteins potentiating the anti-apoptotic function of Bcl-xL.
Organizational Affiliation:
Département de Biologie Structurale Intégrative, Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Université de Strasbourg, CNRS UMR 7104, INSERM U964, 1 rue Laurent Fries, BP 10142, F-67404 Illkirch, France.