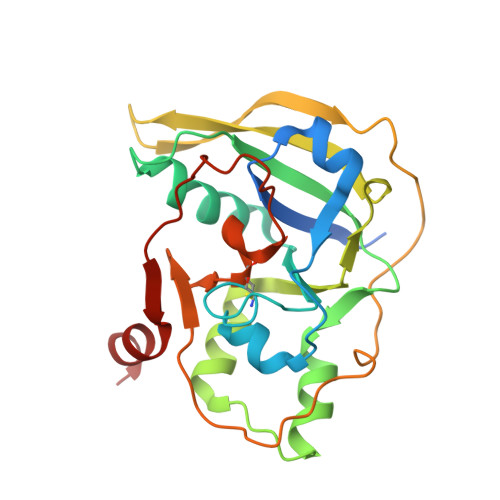
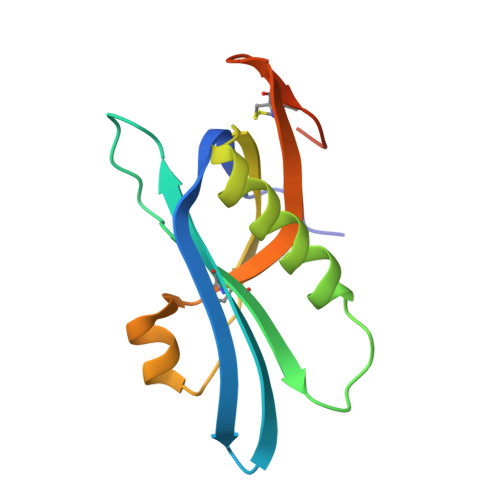
Structure and function analyses of a pertussis-like toxin from pathogenic Escherichia coli reveal a distinct mechanism of inhibition of trimeric G proteins.
Littler, D.R., Ang, S.Y., Moriel, D.G., Kocan, M., Kleifeld, O., Johnson, M.D., Tran, M.T., Paton, A.W., Paton, J.C., Summers, R., Schrembri, M., Rossjohn, J., Beddoe, T.T.(2017) J Biol Chem
- PubMed: 28663369
- DOI: https://doi.org/10.1074/jbc.M117.796094
- Primary Citation of Related Structures:
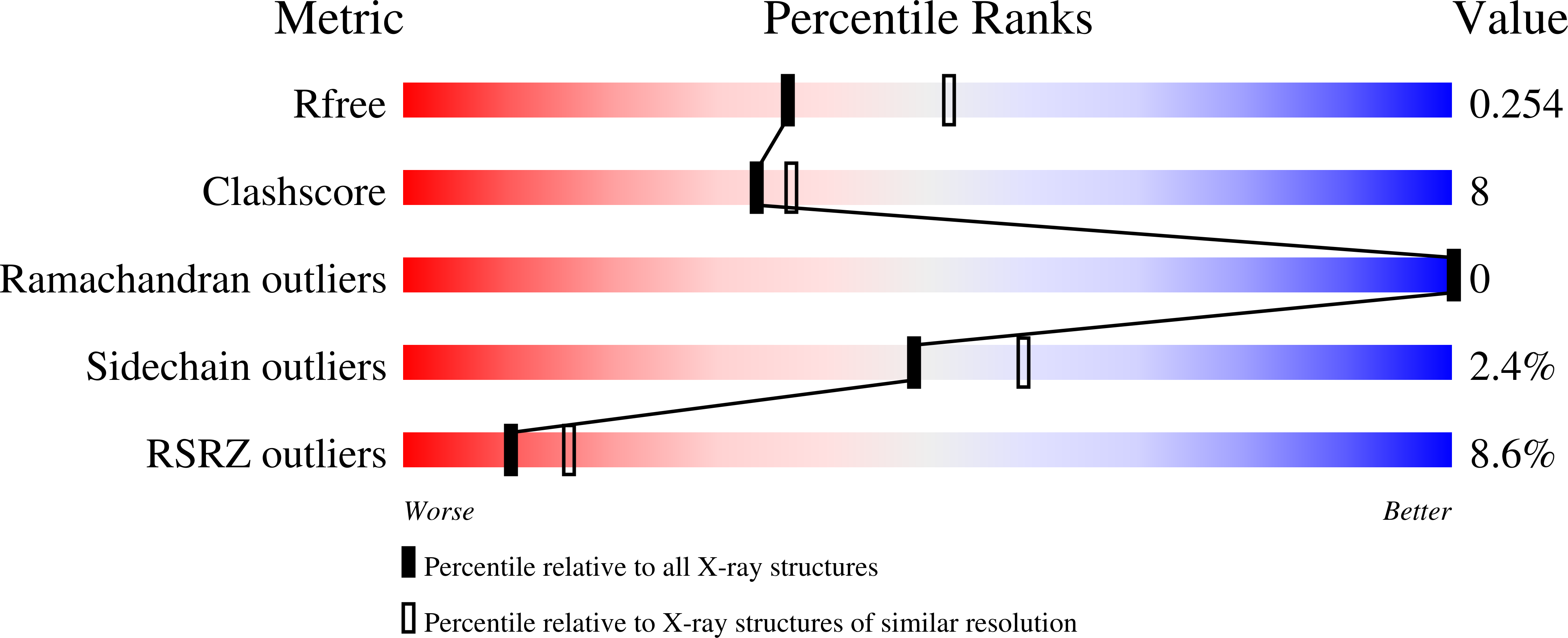
4Z9C, 4Z9D - PubMed Abstract:
Pertussis-like toxins are secreted by several bacterial pathogens during infection. They belong to the AB 5 virulence factors, which bind to glycans on host cell membranes for internalization. Host cell recognition and internalization are mediated by toxin B subunits sharing a unique pentameric ring-like assembly. Although the role of pertussis toxin in whooping cough is well-established, pertussis-like toxins produced by other bacteria are less studied, and their mechanisms of action are unclear. Here, we report that some extra-intestinal Escherichia coli pathogens ( i.e. those that reside in the gut but can spread to other bodily locations) encode a pertussis-like toxin that inhibits mammalian cell growth in vitro We found that this protein, Ec Plt, is related to toxins produced by both nontyphoidal and typhoidal Salmonella serovars. Pertussis-like toxins are secreted as disulfide-bonded heterohexamers in which the catalytic ADP-ribosyltransferase subunit is activated when exposed to the reducing environment in mammalian cells. We found here that the reduced Ec Plt exhibits large structural rearrangements associated with its activation. We noted that inhibitory residues tethered within the NAD + -binding site by an intramolecular disulfide in the oxidized state dissociate upon the reduction and enable loop restructuring to form the nucleotide-binding site. Surprisingly, although pertussis toxin targets a cysteine residue within the α subunit of inhibitory trimeric G-proteins, we observed that activated Ec Plt toxin modifies a proximal lysine/asparagine residue instead. In conclusion, our results reveal the molecular mechanism underpinning activation of pertussis-like toxins, and we also identified differences in host target specificity.
Organizational Affiliation:
From the Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria 3800, Australia.