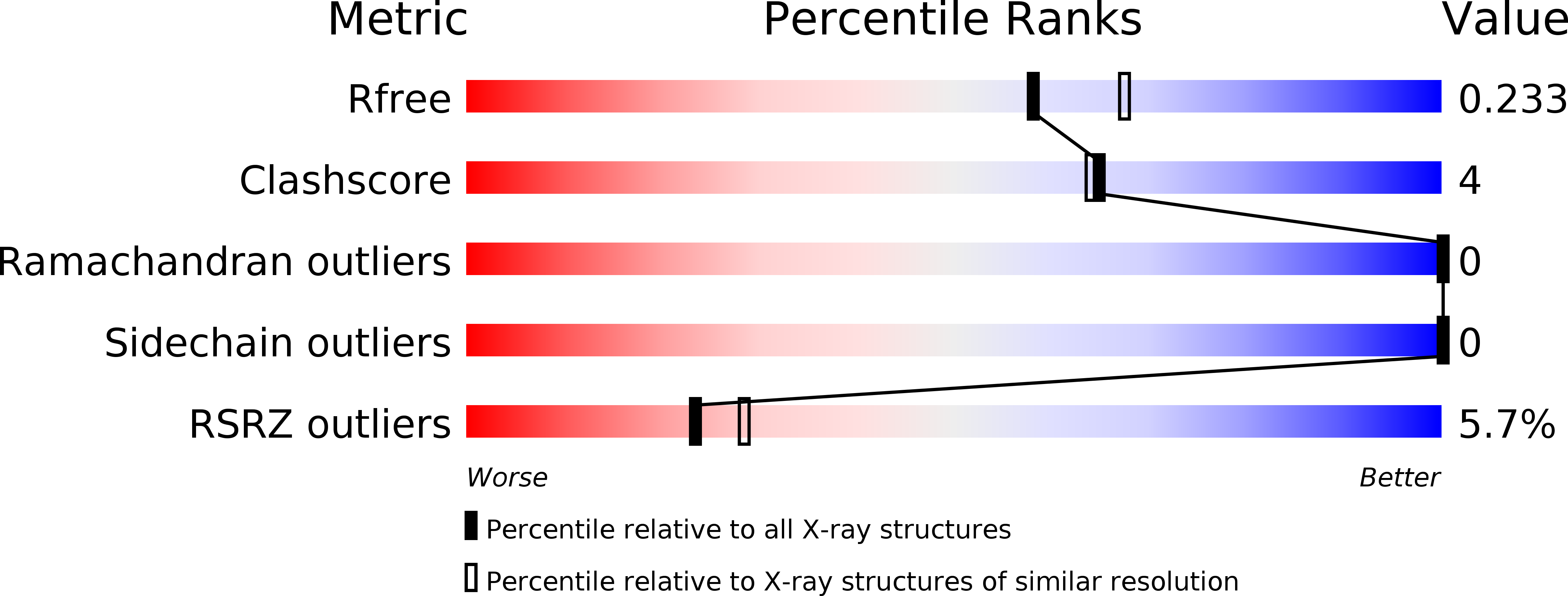
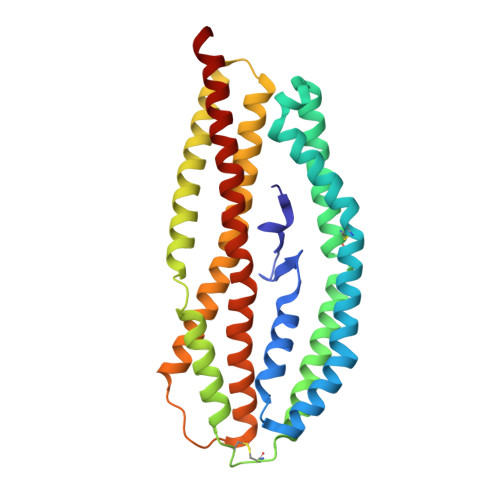
Structurally conserved erythrocyte-binding domain in Plasmodium provides a versatile scaffold for alternate receptor engagement.
Gruszczyk, J., Lim, N.T., Arnott, A., He, W.Q., Nguitragool, W., Roobsoong, W., Mok, Y.F., Murphy, J.M., Smith, K.R., Lee, S., Bahlo, M., Mueller, I., Barry, A.E., Tham, W.H.(2016) Proc Natl Acad Sci U S A 113: E191-E200
- PubMed: 26715754
- DOI: https://doi.org/10.1073/pnas.1516512113
- Primary Citation of Related Structures:
4Z8N - PubMed Abstract:
Understanding how malaria parasites gain entry into human red blood cells is essential for developing strategies to stop blood stage infection. Plasmodium vivax preferentially invades reticulocytes, which are immature red blood cells. The organism has two erythrocyte-binding protein families: namely, the Duffy-binding protein (PvDBP) and the reticulocyte-binding protein (PvRBP) families. Several members of the PvRBP family bind reticulocytes, specifically suggesting a role in mediating host cell selectivity of P. vivax. Here, we present, to our knowledge, the first high-resolution crystal structure of an erythrocyte-binding domain from PvRBP2a, solved at 2.12 Å resolution. The monomeric molecule consists of 10 α-helices and one short β-hairpin, and, although the structural fold is similar to that of PfRh5--the essential invasion ligand in Plasmodium falciparum--its surface properties are distinct and provide a possible mechanism for recognition of alternate receptors. Sequence alignments of the crystallized fragment of PvRBP2a with other PvRBPs highlight the conserved placement of disulfide bonds. PvRBP2a binds mature red blood cells through recognition of an erythrocyte receptor that is neuraminidase- and chymotrypsin-resistant but trypsin-sensitive. By examining the patterns of sequence diversity within field isolates, we have identified and mapped polymorphic residues to the PvRBP2a structure. Using mutagenesis, we have also defined the critical residues required for erythrocyte binding. Characterization of the structural features that govern functional erythrocyte binding for the PvRBP family provides a framework for generating new tools that block P. vivax blood stage infection.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, Parkville, VIC 3052, Australia;