The Non-canonical Tetratricopeptide Repeat (TPR) Domain of Fluorescent (FLU) Mediates Complex Formation with Glutamyl-tRNA Reductase.
Zhang, M., Zhang, F., Fang, Y., Chen, X., Chen, Y., Zhang, W., Dai, H.E., Lin, R., Liu, L.(2015) J Biol Chem 290: 17559-17565
- PubMed: 26037924
- DOI: https://doi.org/10.1074/jbc.M115.662981
- Primary Citation of Related Structures:
4YVO, 4YVQ - PubMed Abstract:
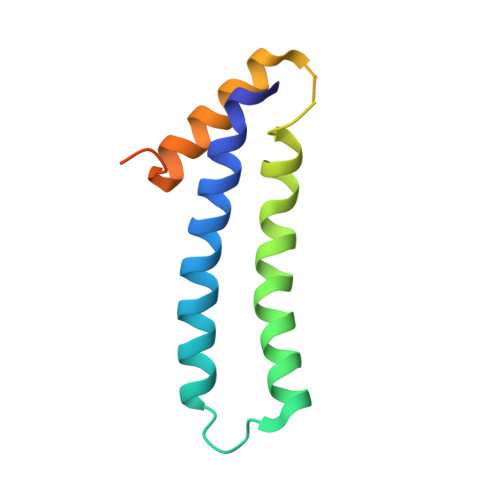
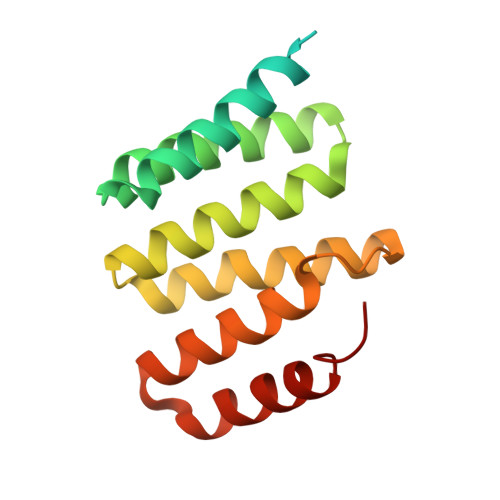
The tetratricopeptide repeat (TPR)-containing protein FLU is a negative regulator of chlorophyll biosynthesis in plants. It directly interacts through its TPR domain with glutamyl-tRNA reductase (GluTR), the rate-limiting enzyme in the formation of δ-aminolevulinic acid (ALA). Delineation of how FLU binds to GluTR is important for understanding the molecular basis for FLU-mediated repression of synthesis of ALA, the universal tetrapyrrole precursor. Here, we characterize the FLU-GluTR interaction by solving the crystal structures of the uncomplexed TPR domain of FLU (FLU(TPR)) at 1.45-Å resolution and the complex of the dimeric domain of GluTR bound to FLU(TPR) at 2.4-Å resolution. Three non-canonical TPR motifs of each FLU(TPR) form a concave surface and clamp the helix bundle in the C-terminal dimeric domain of GluTR. We demonstrate that a 2:2 FLU(TPR)-GluTR complex is the functional unit for FLU-mediated GluTR regulation and suggest that the formation of the FLU-GluTR complex prevents glutamyl-tRNA, the GluTR substrate, from binding with this enzyme. These results also provide insights into the spatial regulation of ALA synthesis by the membrane-located FLU protein.
Organizational Affiliation:
From the School of Life Sciences, Anhui University, Hefei, Anhui 230601.