Synthesis, Biological Activities, and X-ray Crystal Structural Analysis of 25-Hydroxy-25(or 26)-adamantyl-17-[20(22),23-diynyl]-21-norvitamin D Compounds
Watarai, Y., Ishizawa, M., Ikura, T., Zacconi, F.C., Uno, S., Ito, N., Mourino, A., Tokiwa, H., Makishima, M., Yamada, S.(2015) J Med Chem 58: 9510-9521
- PubMed: 26613420
- DOI: https://doi.org/10.1021/acs.jmedchem.5b00792
- Primary Citation of Related Structures:
4YNK - PubMed Abstract:
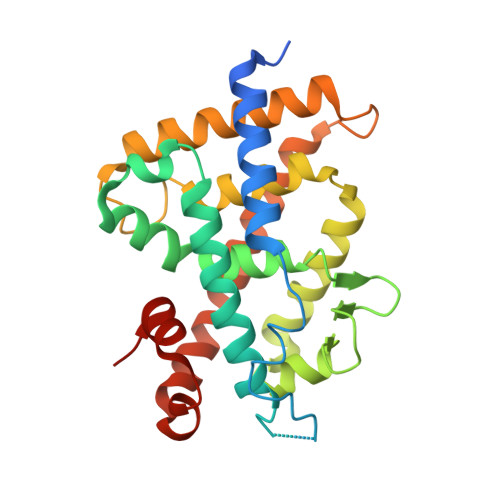
Novel 19-norvitamin D analogues (ADYW1-4, 5a-d) in which an adamantyl diyne side chain is attached directly to the 17-position of the D ring are designed and stereoselectively synthesized. The adamantane ring of these analogues was expected to interfere with helix 12 (H12, activation function 2) of the vitamin D receptor (VDR) to modulate its activities. The analogue 5b binds to the VDR (7% of the natural hormone) and shows significant partial agonistic activity in transactivation assay. Compound 5b showed considerable selectivity in VDR target genes expressions in vitro, it was taken up by target cells 2-3 times more readily, and its lifetime was three times longer than the natural hormone. The X-ray crystal structure of 5b in complex with VDR reveals that the ligand binds similarly to the natural hormone, but the diyne moiety is slightly bent (angles around the diyne 5° to 8°) with respect to the original diyne vitamin D compound 6 in VDR (<1°) due to steric hindrance with helix 12.
Organizational Affiliation:
Department of Chemistry, Faculty of Science, Rikkyo University , Toshima-ku, Tokyo 171-8501, Japan.