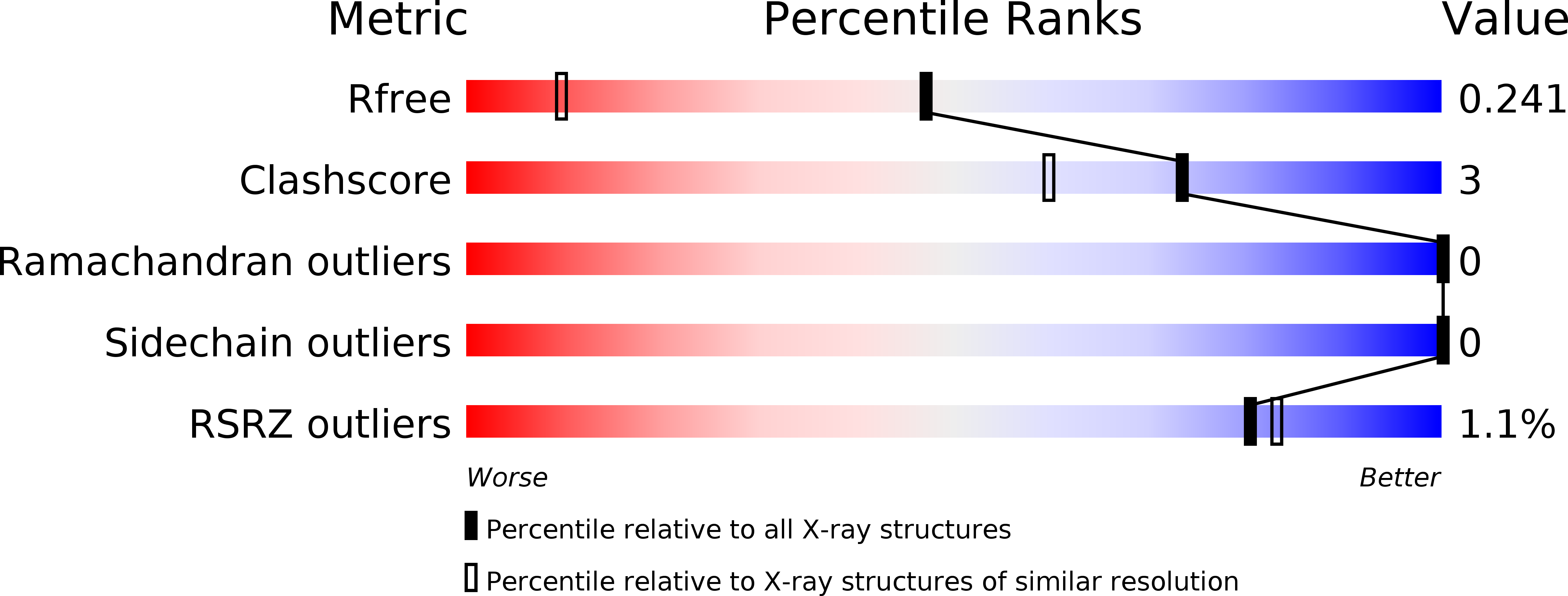
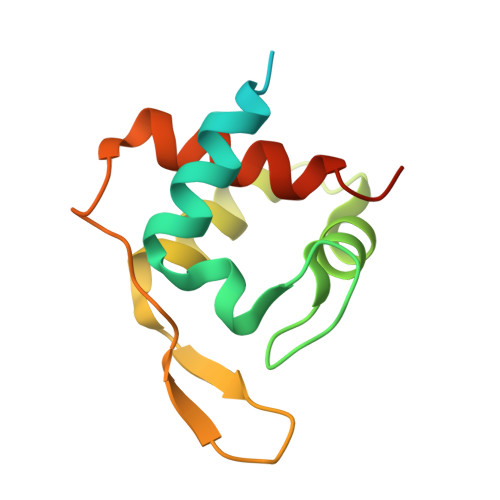
Characterization of MtoD from Sideroxydans lithotrophicus: a cytochrome c electron shuttle used in lithoautotrophic growth.
Beckwith, C.R., Edwards, M.J., Lawes, M., Shi, L., Butt, J.N., Richardson, D.J., Clarke, T.A.(2015) Front Microbiol 6: 332-332
- PubMed: 25972843
- DOI: https://doi.org/10.3389/fmicb.2015.00332
- Primary Citation of Related Structures:
4XXL - PubMed Abstract:
The autotrophic Sideroxydans lithotrophicus ES-1 can grow by coupling the oxidation of ferrous iron to the reduction of oxygen. Soluble ferrous iron is oxidized at the surface of the cell by an MtoAB porin-cytochrome complex that functions as an electron conduit through the outer membrane. Electrons are then transported to the cytoplasmic membrane where they are used to generate proton motive force (PMF) (for ATP synthesis) and NADH for autotrophic processes such as carbon fixation. As part of the mtoAB gene cluster, S. lithotrophicus also contains the gene mtoD that is proposed to encode a cytochrome c protein. We isolated mtoD from a Shewanella oneidensis expression system where the mtoD gene was expressed on a pBAD plasmid vector. Biochemical, biophysical, and crystallographic characterization of the purified MtoD revealed it as an 11 kDa monomeric protein containing a single heme. Sequence and structural alignment indicated that MtoD belonged to the class-1 cytochrome c family and had a similar fold to ferricytochrome c552 family, however the MtoD heme is bis-histidine coordinated and is substantially more exposed than the hemes of other family members. The reduction potential of the MtoD heme at pH 7 was +155 mV vs. Standard Hydrogen Electrode, which is approximately 100 mV lower than that of mitochondrial cytochrome c. Consideration of the properties of MtoD in the context of the potential respiratory partners identified from the genome suggests that MtoD could associate to multiple electron transfer partners as the primary periplasmic electron shuttle.
Organizational Affiliation:
Centre for Molecular and Structural Biochemistry, School of Biological Sciences and School of Chemistry, University of East Anglia Norwich, UK.