Biochemical and structural characterization of MUPP1-PDZ4 domain from Mus musculus.
Zhu, H., Liu, Z., Huang, Y., Zhang, C., Li, G., Liu, W.(2015) Acta Biochim Biophys Sin (Shanghai) 47: 199-206
- PubMed: 25662616
- DOI: https://doi.org/10.1093/abbs/gmv002
- Primary Citation of Related Structures:
4XH7 - PubMed Abstract:
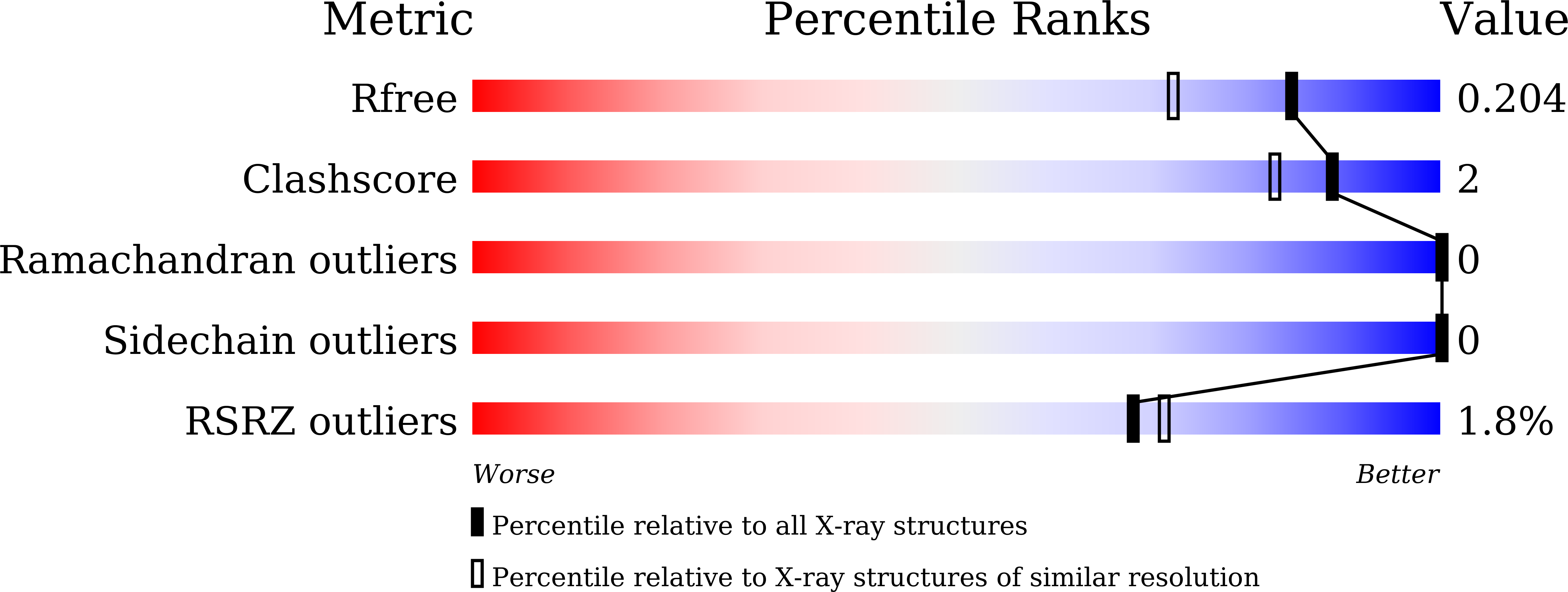
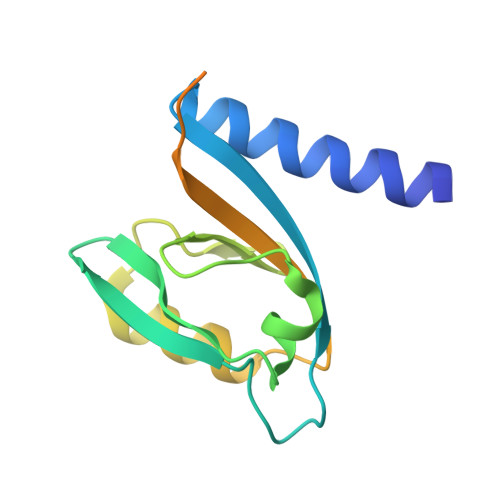
Specific protein-protein interactions are important for biological signal transduction. The postsynaptic density-95, disc-large, and zonulin-1 (PDZ) domain is one of the most abundant protein interaction modules. Multi-PDZ-domain protein 1 (MUPP1), as a scaffold protein, contains 13 PDZ domains and plays an important role in cytoskeletal organization, cell polarity, and cell proliferation. The study on PDZ domain of MUPP1 helps to understand the mechanisms and functions of MUPP1. In the present study, the fourth PDZ domain of MUPP1 (MUPP1-PDZ4) from Mus musculus was cloned, expressed, purified, and characterized. The MUPP1-PDZ4 domain was subcloned into a pET-vector and expressed in Escherichia coli. Affinity chromatography and size-exclusion chromatography were used to purify the protein. MUPP1-PDZ4 protein was a monomer with a molar mass of 16.4 kDa in solution and had a melting point of 60.3°C. Using the sitting-drop vapor-diffusion method, MUPP1-PDZ4 protein crystals were obtained in a solution (pH 7.0) containing 2% (v/v) polyethylene glycol 400, 0.1 M imidazole, and 24% (w/v) polyethylene glycol monoethyl ether 5000. Finally, the crystal was diffracted with 1.6 Å resolution. The crystal structure showed that MUPP1-PDZ4 domain contained three α-helices and six β-strands in the core. The GLGI motif, L562/A564 on the β-strand B, and H605/V608/L612 on the α-helix B formed a PDZ binding pocket which could bind to the C-terminal of the binding partners. This biochemical and structural information will provide insights into how PDZ binds to its target peptide and the theoretical foundation for the function of MUPP1.
Organizational Affiliation:
Shenzhen Key Laboratory for Neuronal Structural Biology, Biomedical Research Institute, Shenzhen Peking University-The Hong Kong University of Science and Technology Medical Center, Shenzhen 518036, China.