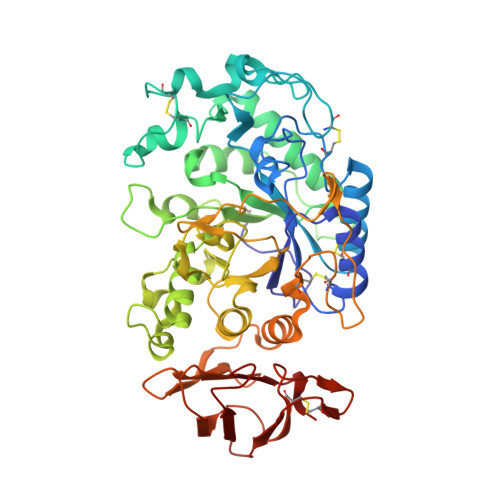
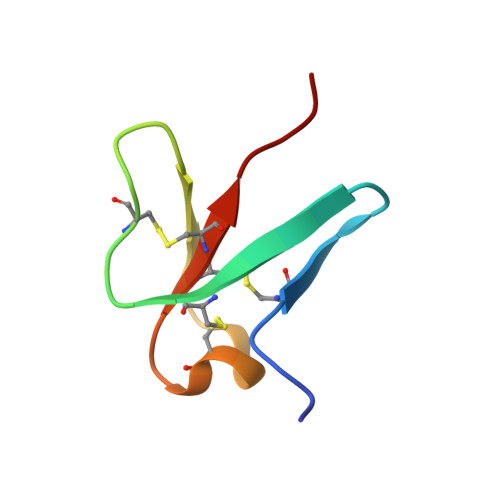
Structural Templating and Guided Refolding of the Potent Naturally Occurring Peptide Helianthamide Within the Active Site of Amylase, a Diabetes and Obesity Therapeutic Target
Tysoe, C., Williams, L.K., Keyzers, R., Nguyen, N.T., Aguda, A., Zhang, X., Tarling, C.A., Andersen, R.J., Brayer, G.D., Withers, S.G.To be published.