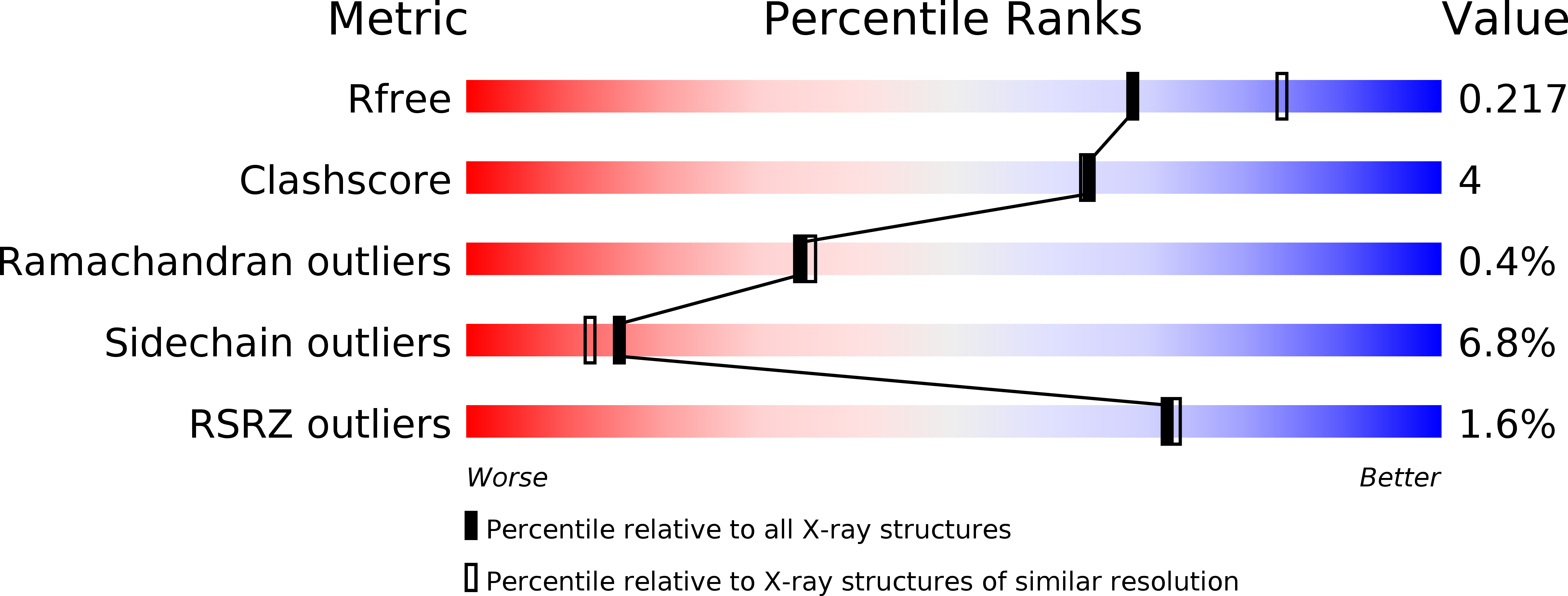
Decreasing the Rate of Metabolic Ketone Reduction in the Discovery of a Clinical Acetyl-CoA Carboxylase Inhibitor for the Treatment of Diabetes.
Griffith, D.A., Kung, D.W., Esler, W.P., Amor, P.A., Bagley, S.W., Beysen, C., Carvajal-Gonzalez, S., Doran, S.D., Limberakis, C., Mathiowetz, A.M., McPherson, K., Price, D.A., Ravussin, E., Sonnenberg, G.E., Southers, J.A., Sweet, L.J., Turner, S.M., Vajdos, F.F.(2014) J Med Chem 57: 10512-10526
- PubMed: 25423286
- DOI: https://doi.org/10.1021/jm5016022
- Primary Citation of Related Structures:
4WYO, 4WZ8 - PubMed Abstract:
Acetyl-CoA carboxylase (ACC) inhibitors offer significant potential for the treatment of type 2 diabetes mellitus (T2DM), hepatic steatosis, and cancer. However, the identification of tool compounds suitable to test the hypothesis in human trials has been challenging. An advanced series of spirocyclic ketone-containing ACC inhibitors recently reported by Pfizer were metabolized in vivo by ketone reduction, which complicated human pharmacology projections. We disclose that this metabolic reduction can be greatly attenuated through introduction of steric hindrance adjacent to the ketone carbonyl. Incorporation of weakly basic functionality improved solubility and led to the identification of 9 as a clinical candidate for the treatment of T2DM. Phase I clinical studies demonstrated dose-proportional increases in exposure, single-dose inhibition of de novo lipogenesis (DNL), and changes in indirect calorimetry consistent with increased whole-body fatty acid oxidation. This demonstration of target engagement validates the use of compound 9 to evaluate the role of DNL in human disease.
Organizational Affiliation:
Worldwide Medicinal Chemistry, ‡Cardiovascular, Metabolic and Endocrine Diseases Research Unit, and ∥Clinical Research Statistics, Pfizer Worldwide Research and Development , Cambridge, Massachusetts 02139, United States.