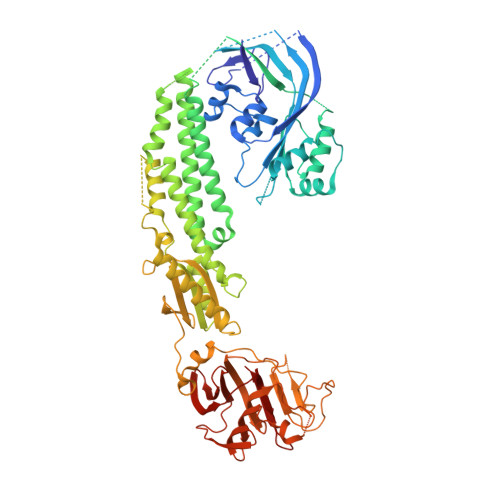
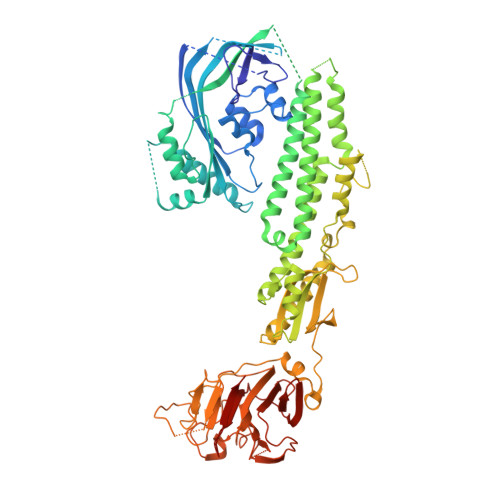
Stonefish toxin defines an ancient branch of the perforin-like superfamily.
Ellisdon, A.M., Reboul, C.F., Panjikar, S., Huynh, K., Oellig, C.A., Winter, K.L., Dunstone, M.A., Hodgson, W.C., Seymour, J., Dearden, P.K., Tweten, R.K., Whisstock, J.C., McGowan, S.(2015) Proc Natl Acad Sci U S A 112: 15360-15365
- PubMed: 26627714
- DOI: https://doi.org/10.1073/pnas.1507622112
- Primary Citation of Related Structures:
4WVM - PubMed Abstract:
The lethal factor in stonefish venom is stonustoxin (SNTX), a heterodimeric cytolytic protein that induces cardiovascular collapse in humans and native predators. Here, using X-ray crystallography, we make the unexpected finding that SNTX is a pore-forming member of an ancient branch of the Membrane Attack Complex-Perforin/Cholesterol-Dependent Cytolysin (MACPF/CDC) superfamily. SNTX comprises two homologous subunits (α and β), each of which comprises an N-terminal pore-forming MACPF/CDC domain, a central focal adhesion-targeting domain, a thioredoxin domain, and a C-terminal tripartite motif family-like PRY SPla and the RYanodine Receptor immune recognition domain. Crucially, the structure reveals that the two MACPF domains are in complex with one another and arranged into a stable early prepore-like assembly. These data provide long sought after near-atomic resolution insights into how MACPF/CDC proteins assemble into prepores on the surface of membranes. Furthermore, our analyses reveal that SNTX-like MACPF/CDCs are distributed throughout eukaryotic life and play a broader, possibly immune-related function outside venom.
Organizational Affiliation:
Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Melbourne, VIC, 3800, Australia; Australian Research Council Centre of Excellence in Advanced Molecular Imaging, Monash University, Melbourne, VIC, 3800, Australia;