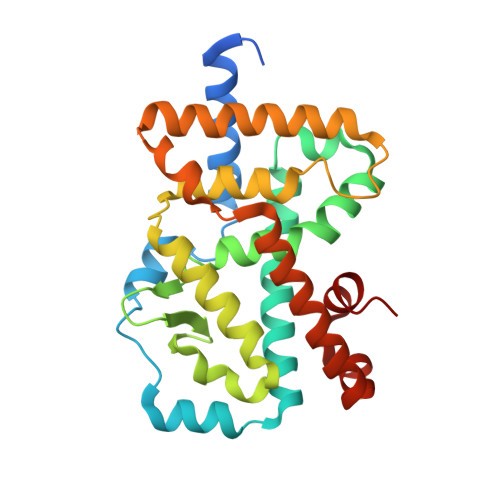
Minor Structural Change to Tertiary Sulfonamide RORc Ligands Led to Opposite Mechanisms of Action.
Rene, O., Fauber, B.P., de Leon Boenig, G., Burton, B., Eidenschenk, C., Everett, C., Gobbi, A., Hymowitz, S.G., Johnson, A.R., Kiefer, J.R., Liimatta, M., Lockey, P., Norman, M., Ouyang, W., Wallweber, H.A., Wong, H.(2015) ACS Med Chem Lett 6: 276-281
- PubMed: 25815138
- DOI: https://doi.org/10.1021/ml500420y
- Primary Citation of Related Structures:
4WPF, 4WQP - PubMed Abstract:
A minor structural change to tertiary sulfonamide RORc ligands led to distinct mechanisms of action. Co-crystal structures of two compounds revealed mechanistically consistent protein conformational changes. Optimized phenylsulfonamides were identified as RORc agonists while benzylsulfonamides exhibited potent inverse agonist activity. Compounds behaving as agonists in our biochemical assay also gave rise to an increased production of IL-17 in human PBMCs whereas inverse agonists led to significant suppression of IL-17 under the same assay conditions. The most potent inverse agonist compound showed >180-fold selectivity over the ROR isoforms as well as all other nuclear receptors that were profiled.
Organizational Affiliation:
Genentech, Inc., 1 DNA Way, South San Francisco, California 94080, United States.