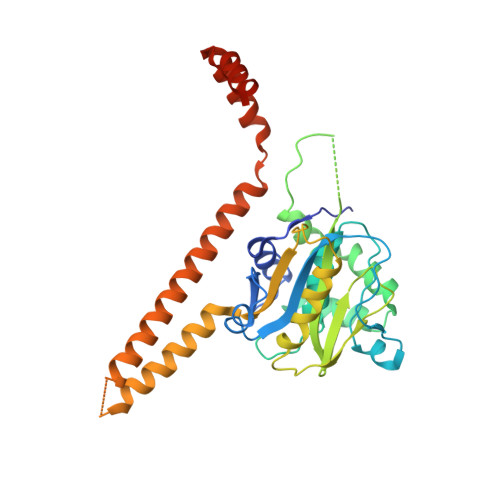
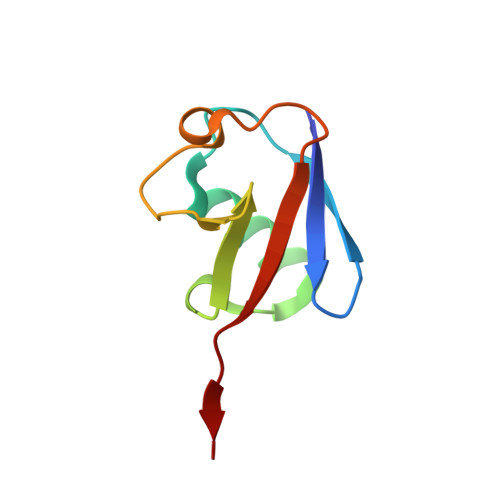
Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
VanderLinden, R.T., Hemmis, C.W., Schmitt, B., Ndoja, A., Whitby, F.G., Robinson, H., Cohen, R.E., Yao, T., Hill, C.P.(2015) Mol Cell 57: 901-911
- PubMed: 25702872
- DOI: https://doi.org/10.1016/j.molcel.2015.01.016
- Primary Citation of Related Structures:
4WLP, 4WLQ, 4WLR - PubMed Abstract:
The UCH37 deubiquitylase functions in two large and very different complexes, the 26S proteasome and the INO80 chromatin remodeler. We have performed biochemical characterization and determined crystal structures of UCH37 in complexes with RPN13 and NFRKB, which mediate its recruitment to the proteasome and INO80, respectively. RPN13 and NFRKB make similar contacts to the UCH37 C-terminal domain but quite different contacts to the catalytic UCH domain. RPN13 can activate UCH37 by disrupting dimerization, although physiologically relevant activation likely results from stabilization of a surface competent for ubiquitin binding and modulation of the active-site crossover loop. In contrast, NFRKB inhibits UCH37 by blocking the ubiquitin-binding site and by disrupting the enzyme active site. These findings reveal remarkable commonality in mechanisms of recruitment, yet very different mechanisms of regulating enzyme activity, and provide a foundation for understanding the roles of UCH37 in the unrelated proteasome and INO80 complexes.
Organizational Affiliation:
Department of Biochemistry University of Utah School of Medicine, Salt Lake City, UT 84112-5650 USA.