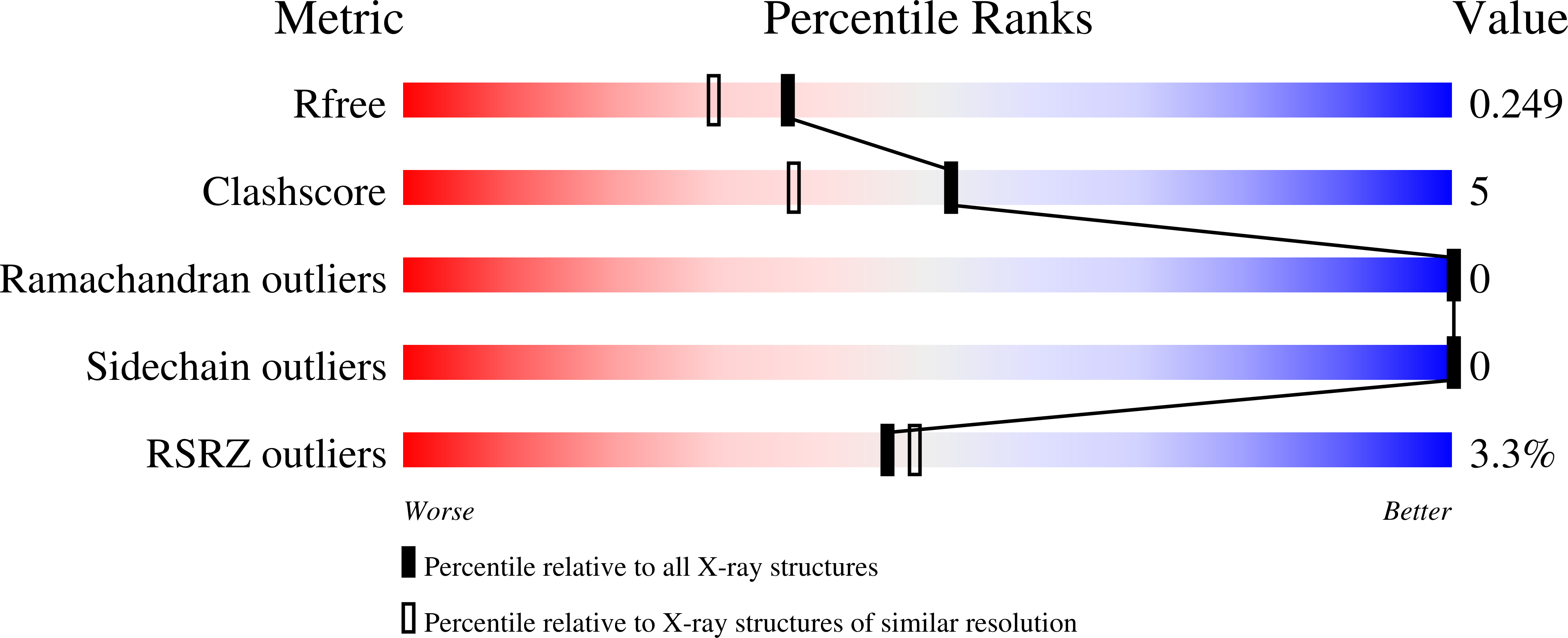
c-di-AMP recognition by Staphylococcus aureus PstA.
Muller, M., Hopfner, K., Witte, G.(2015) FEBS Lett 589: 45-51
- PubMed: 25435171
- DOI: https://doi.org/10.1016/j.febslet.2014.11.022
- Primary Citation of Related Structures:
4WK1, 4WK3 - PubMed Abstract:
Cyclic-di-AMP (c-di-AMP) is a bacterial secondary messenger involved in various processes, including sensing of DNA-integrity, cell wall metabolism and potassium transport. A number of c-di-AMP receptor proteins have recently been identified in Staphylococcus aureus. One of them - PstA - possesses a ferredoxin-like fold and is structurally related to the class of PII signal-transduction proteins. PII proteins are involved in a large number of pathways, most of them associated with nitrogen metabolism. In this study we describe the mode of c-di-AMP binding and subsequent structural changes of S. aureus PstA. An altered architecture in PstA compared to canonical PII proteins results in differences in ligand coordination.
Organizational Affiliation:
Ludwig-Maximilians-Universität München, Gene Center and Dept. of Biochemistry, Feodor-Lynen-Str. 25, 81377 Munich, Germany.