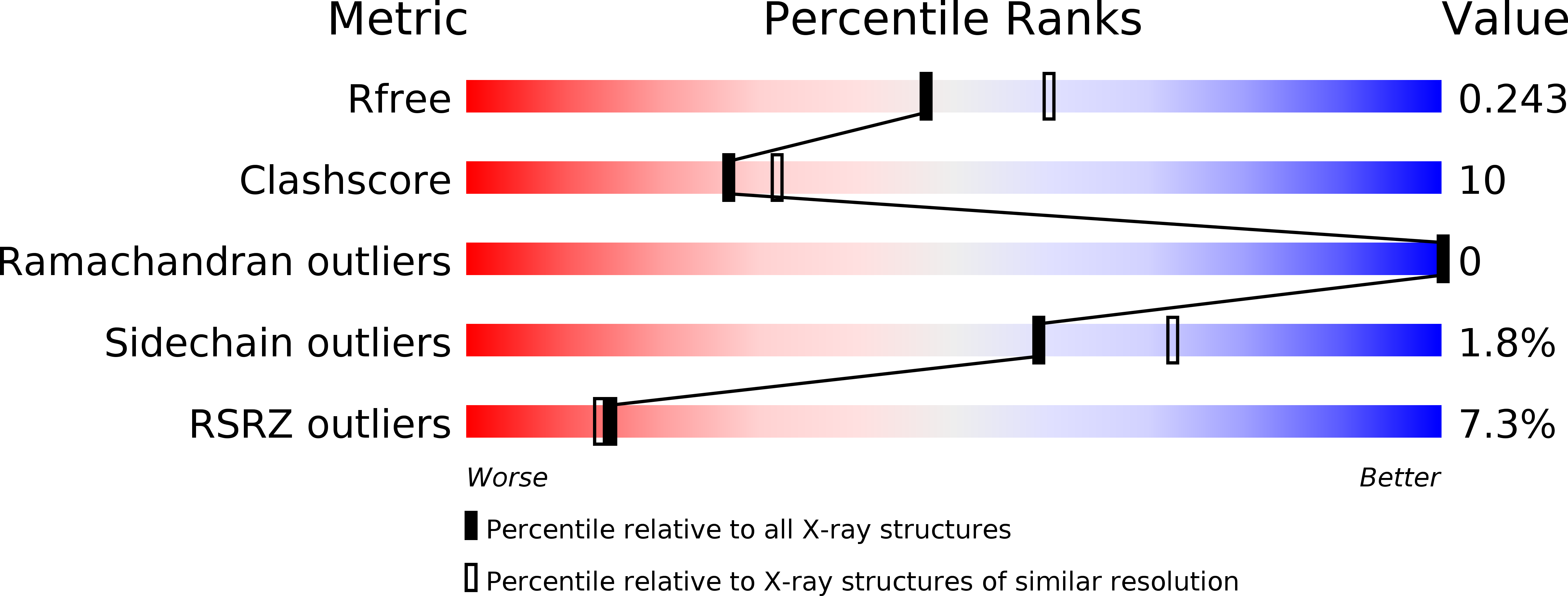
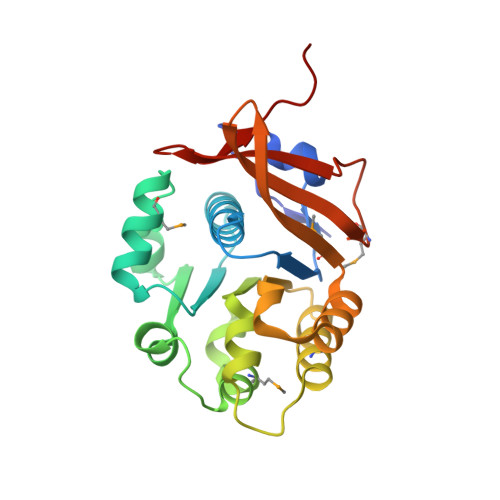
Crystal structure of the flagellar accessory protein FlaH of Methanocaldococcus jannaschii suggests a regulatory role in archaeal flagellum assembly.
Meshcheryakov, V.A., Wolf, M.(2016) Protein Sci 25: 1147-1155
- PubMed: 27060465
- DOI: https://doi.org/10.1002/pro.2932
- Primary Citation of Related Structures:
4WIA - PubMed Abstract:
Archaeal flagella are unique structures that share functional similarity with bacterial flagella, but are structurally related to bacterial type IV pili. The flagellar accessory protein FlaH is one of the conserved components of the archaeal motility system. However, its function is not clearly understood. Here, we present the 2.2 Å resolution crystal structure of FlaH from the hyperthermophilic archaeon, Methanocaldococcus jannaschii. The protein has a characteristic RecA-like fold, which has been found previously both in archaea and bacteria. We show that FlaH binds to immobilized ATP-however, it lacks ATPase activity. Surface plasmon resonance analysis demonstrates that ATP affects the interaction between FlaH and the archaeal motor protein FlaI. In the presence of ATP, the FlaH-FlaI interaction becomes significantly weaker. A database search revealed similarity between FlaH and several DNA-binding proteins of the RecA superfamily. The closest structural homologs of FlaH are KaiC-like proteins, which are archaeal homologs of the circadian clock protein KaiC from cyanobacteria. We propose that one of the functions of FlaH may be the regulation of archaeal motor complex assembly.
Organizational Affiliation:
Molecular Cryo-Electron Microscopy Unit, Okinawa Institute of Science and Technology Graduate University, 1919-1 Tancha, Onna, Kunigami, Okinawa 904-0495, Japan.