Identification and characterization of amlexanox as a g protein-coupled receptor kinase 5 inhibitor.
Homan, K.T., Wu, E., Cannavo, A., Koch, W.J., Tesmer, J.J.(2014) Molecules 19: 16937-16949
- PubMed: 25340299
- DOI: https://doi.org/10.3390/molecules191016937
- Primary Citation of Related Structures:
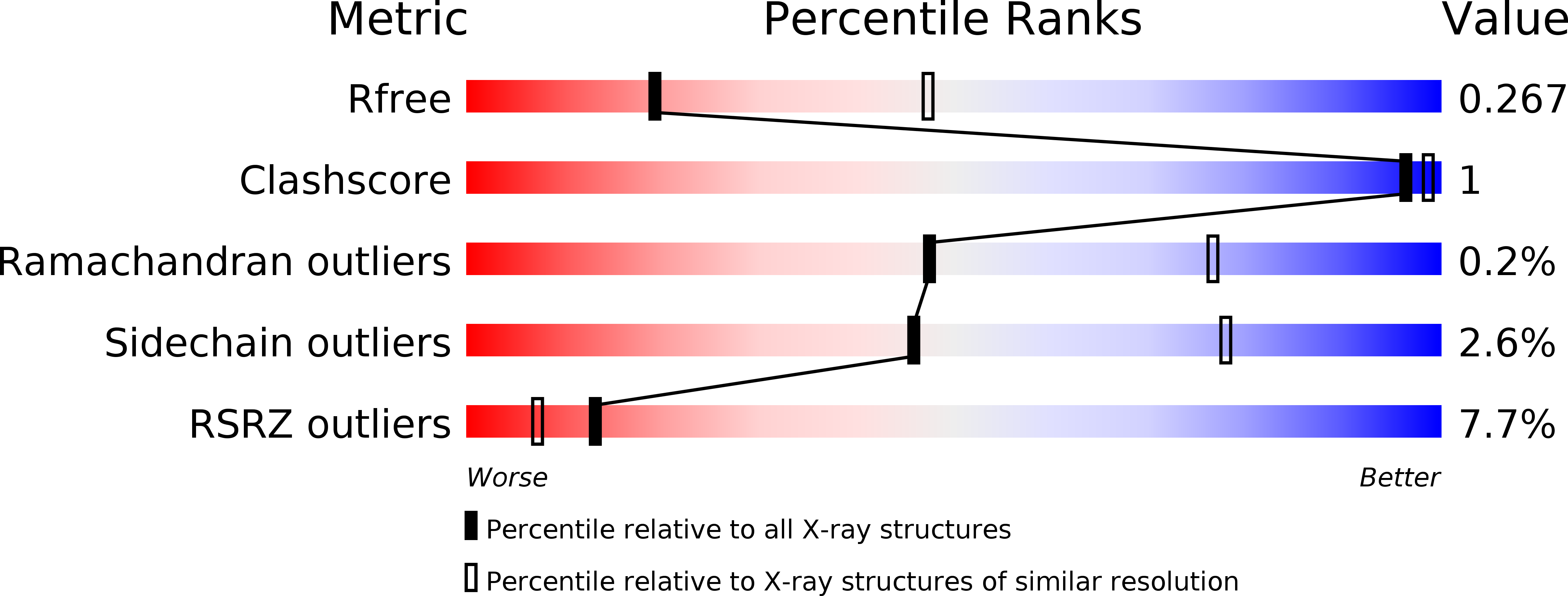
4WBO - PubMed Abstract:
G protein-coupled receptor kinases (GRKs) have been implicated in human diseases ranging from heart failure to diabetes. Previous studies have identified several compounds that selectively inhibit GRK2, such as paroxetine and balanol. Far fewer selective inhibitors have been reported for GRK5, a target for the treatment of cardiac hypertrophy, and the mechanism of action of reported compounds is unknown. To identify novel scaffolds that selectively inhibit GRK5, a differential scanning fluorometry screen was used to probe a library of 4480 compounds. The best hit was amlexanox, an FDA-approved anti-inflammatory, anti-allergic immunomodulator. The crystal structure of amlexanox in complex with GRK1 demonstrates that its tricyclic aromatic ring system forms ATP-like interactions with the hinge of the kinase domain, which is likely similar to how this drug binds to IκB kinase ε (IKKε), another kinase known to be inhibited by this compound. Amlexanox was also able to inhibit myocyte enhancer factor 2 transcriptional activity in neonatal rat ventricular myocytes in a manner consistent with GRK5 inhibition. The GRK1 amlexanox structure thus serves as a springboard for the rational design of inhibitors with improved potency and selectivity for GRK5 and IKKε.
Organizational Affiliation:
Life Sciences Institute and the Departments of Pharmacology and Biological Sciences, University of Michigan, Ann Arbor, MI 48109, USA.