Phosphorylation of Synaptic Vesicle Protein 2A at Thr84 by Casein Kinase 1 Family Kinases Controls the Specific Retrieval of Synaptotagmin-1.
Zhang, N., Gordon, S.L., Fritsch, M.J., Esoof, N., Campbell, D.G., Gourlay, R., Velupillai, S., Macartney, T., Peggie, M., Van Aalten, D.M.F., Cousin, M.A., Alessi, D.R.(2015) J Neurosci 35: 2492
- PubMed: 25673844
- DOI: https://doi.org/10.1523/JNEUROSCI.4248-14.2015
- Primary Citation of Related Structures:
4V11 - PubMed Abstract:
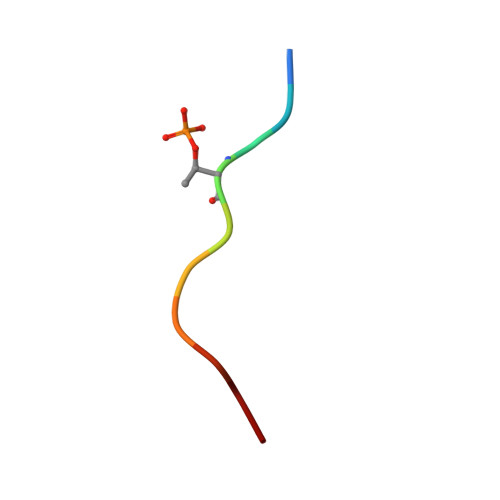
Synaptic vesicle protein 2A (SV2A) is a ubiquitous component of synaptic vesicles (SVs). It has roles in both SV trafficking and neurotransmitter release. We demonstrate that Casein kinase 1 family members, including isoforms of Tau-tubulin protein kinases (TTBK1 and TTBK2), phosphorylate human SV2A at two constellations of residues, namely Cluster-1 (Ser42, Ser45, and Ser47) and Cluster-2 (Ser80, Ser81, and Thr84). These residues are also phosphorylated in vivo, and the phosphorylation of Thr84 within Cluster-2 is essential for triggering binding to the C2B domain of human synaptotagmin-1. We show by crystallographic and other analyses that the phosphorylated Thr84 residue binds to a pocket formed by three conserved Lys residues (Lys314, Lys326, and Lys328) on the surface of the synaptotagmin-1 C2B domain. Finally, we observed dysfunctional synaptotagmin-1 retrieval during SV endocytosis by ablating its phospho-dependent interaction with SV2A, knockdown of SV2A, or rescue with a phosphorylation-null Thr84 SV2A mutant in primary cultures of mouse neurons. This study reveals fundamental details of how phosphorylation of Thr84 on SV2A controls its interaction with synaptotagmin-1 and implicates SV2A as a phospho-dependent chaperone required for the specific retrieval of synaptotagmin-1 during SV endocytosis.
Organizational Affiliation:
Medical Research Council Protein Phosphorylation and Ubiquitylation Unit, College of Life Sciences, University of Dundee, Dundee DD1 5EH, Scotland, United Kingdom, and.