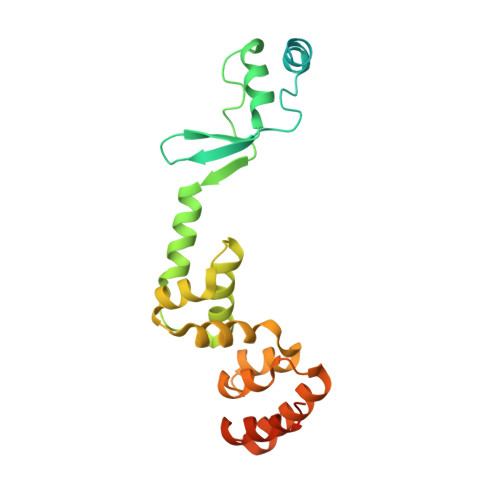
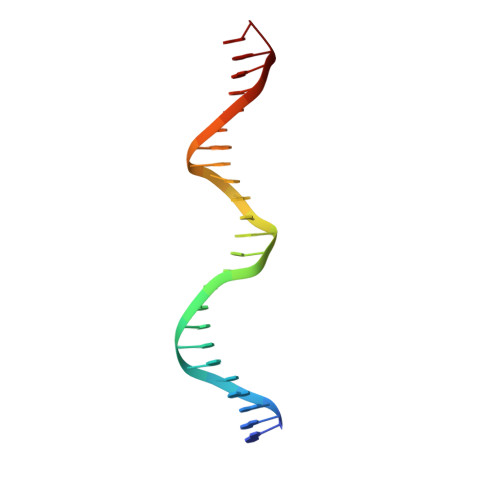
Insights into ParB spreading from the complex structure of Spo0J and parS.
Chen, B.W., Lin, M.H., Chu, C.H., Hsu, C.E., Sun, Y.J.(2015) Proc Natl Acad Sci U S A 112: 6613-6618
- PubMed: 25964325
- DOI: https://doi.org/10.1073/pnas.1421927112
- Primary Citation of Related Structures:
4UMK - PubMed Abstract:
Spo0J (stage 0 sporulation protein J, a member of the ParB superfamily) is an essential component of the ParABS (partition system of ParA, ParB, and parS)-related bacterial chromosome segregation system. ParB (partition protein B) and its regulatory protein, ParA, act cooperatively through parS (partition S) DNA to facilitate chromosome segregation. ParB binds to chromosomal DNA at specific parS sites as well as the neighboring nonspecific DNA sites. Various ParB molecules can associate together and spread along the chromosomal DNA. ParB oligomer and parS DNA interact together to form a high-order nucleoprotein that is required for the loading of the structural maintenance of chromosomes proteins onto the chromosome for chromosomal DNA condensation. In this report, we characterized the binding of parS and Spo0J from Helicobacter pylori (HpSpo0J) and solved the crystal structure of the C-terminal domain truncated protein (Ct-HpSpo0J)-parS complex. Ct-HpSpo0J folds into an elongated structure that includes a flexible N-terminal domain for protein-protein interaction and a conserved DNA-binding domain for parS binding. Two Ct-HpSpo0J molecules bind with one parS. Ct-HpSpo0J interacts vertically and horizontally with its neighbors through the N-terminal domain to form an oligomer. These adjacent and transverse interactions are accomplished via a highly conserved arginine patch: RRLR. These interactions might be needed for molecular assembly of a high-order nucleoprotein complex and for ParB spreading. A structural model for ParB spreading and chromosomal DNA condensation that lead to chromosome segregation is proposed.
Organizational Affiliation:
Department of Life Science and Institute of Bioinformatics and Structural Biology, College of Life Science, National Tsing Hua University, Hsinchu 30013, Taiwan.