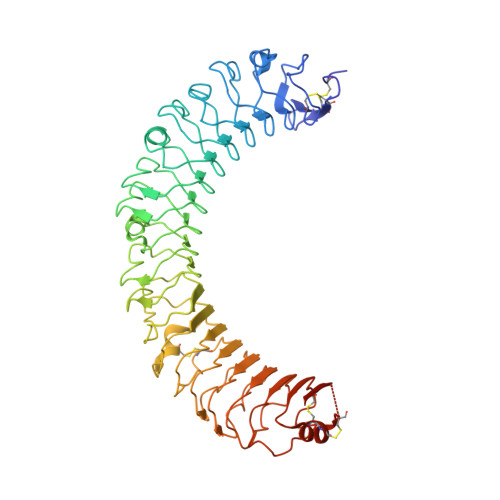
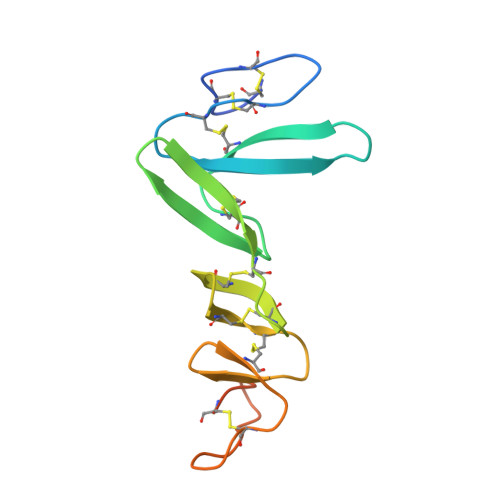
Crystal Structure of R-Spondin 2 in Complex with the Ectodomains of its Receptors Lgr5 and Znrf3.
Zebisch, M., Jones, E.Y.(2015) J Struct Biol 191: 149
- PubMed: 26123262
- DOI: https://doi.org/10.1016/j.jsb.2015.05.008
- Primary Citation of Related Structures:
4UFR, 4UFS - PubMed Abstract:
The four secreted R-spondin (Rspo1-4) proteins of vertebrates function as stem cell growth factors and potentiate canonical Wnt signalling. Rspo proteins act by cross-linking members of two cell surface receptor families, complexing the stem cell markers LGR4-6 with the Frizzled-specific E3 ubiquitin ligases ZNRF3/RNF43. The consequent internalisation of the ternary LGR-Rspo-E3 complex removes the E3 ligase activity, which otherwise targets the Wnt receptor Frizzled for degradation, and thus enhances Wnt signalling. Multiple combinations of LGR4-6, Rspo1-4 and ZNRF3/RNF43 are possible, implying the existence of generic interaction determinants, but also of specific differences in complex architecture and activity. We present here a high resolution crystal structure of an ectodomain variant of human LGR5 (hLGR5ecto) complexed with a signalling competent fragment of mouse Rspo2 (mRspo2Fu1-Fu2). The structure shows that the particularly potent Rspo2 ligand engages LGR5 in a fashion almost identical to that reported for hRSPO1. Comparison of our hLGR5ecto structure with previously published structures highlights a surprising plasticity of the LGR ectodomains, characterised by a nearly 9° or larger rotation of the N-terminal half of the horseshoe-like fold relative to the C-terminal half. We also report a low resolution hLGR5-mRspo2Fu1-Fu2-mZNRF3ecto ternary complex structure. This crystal structure confirms our previously suggested hypothesis, showing that Rspo proteins cross-link LGRs and ZNRF3 into a 2:2:2 complex, whereas a 1:1:1 complex is formed with RNF43.
Organizational Affiliation:
Division of Structural Biology, Henry Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, United Kingdom.