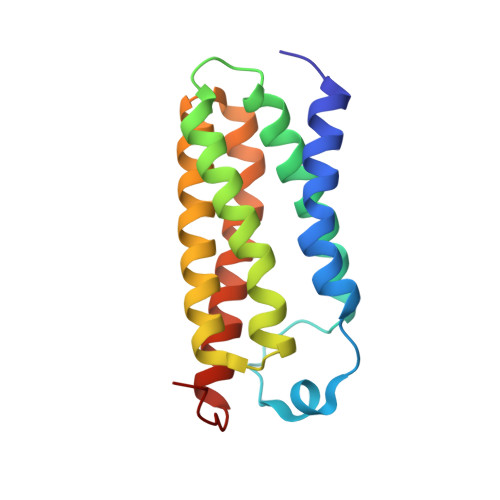
Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Li, F., Liu, J., Zheng, Y., Garavito, R.M., Ferguson-Miller, S.(2015) Science 347: 555-558
- PubMed: 25635101
- DOI: https://doi.org/10.1126/science.1260590
- Primary Citation of Related Structures:
4UC1, 4UC2, 4UC3 - PubMed Abstract:
The 18-kilodalton translocator protein (TSPO), proposed to be a key player in cholesterol transport into mitochondria, is highly expressed in steroidogenic tissues, metastatic cancer, and inflammatory and neurological diseases such as Alzheimer's and Parkinson's. TSPO ligands, including benzodiazepine drugs, are implicated in regulating apoptosis and are extensively used in diagnostic imaging. We report crystal structures (at 1.8, 2.4, and 2.5 angstrom resolution) of TSPO from Rhodobacter sphaeroides and a mutant that mimics the human Ala(147)→Thr(147) polymorphism associated with psychiatric disorders and reduced pregnenolone production. Crystals obtained in the lipidic cubic phase reveal the binding site of an endogenous porphyrin ligand and conformational effects of the mutation. The three crystal structures show the same tightly interacting dimer and provide insights into the controversial physiological role of TSPO and how the mutation affects cholesterol binding.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824, USA.