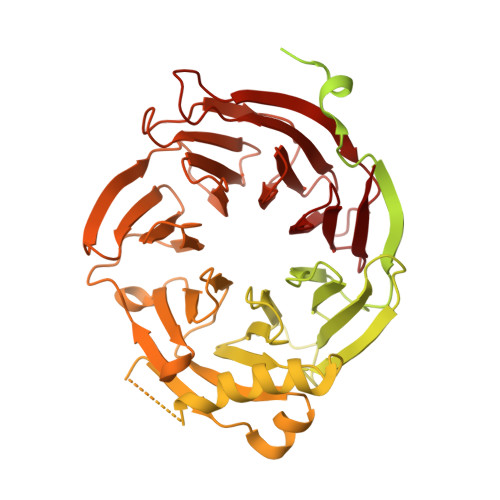
The Carboxy-Terminal Domain of Erb1 Is a Seven-Bladed -Propeller that Binds RNA.
Wegrecki, M., Neira, J.L., Bravo, J.(2015) PLoS One 10: e0123463-e0123463
- PubMed: 25880847
- DOI: https://doi.org/10.1371/journal.pone.0123463
- Primary Citation of Related Structures:
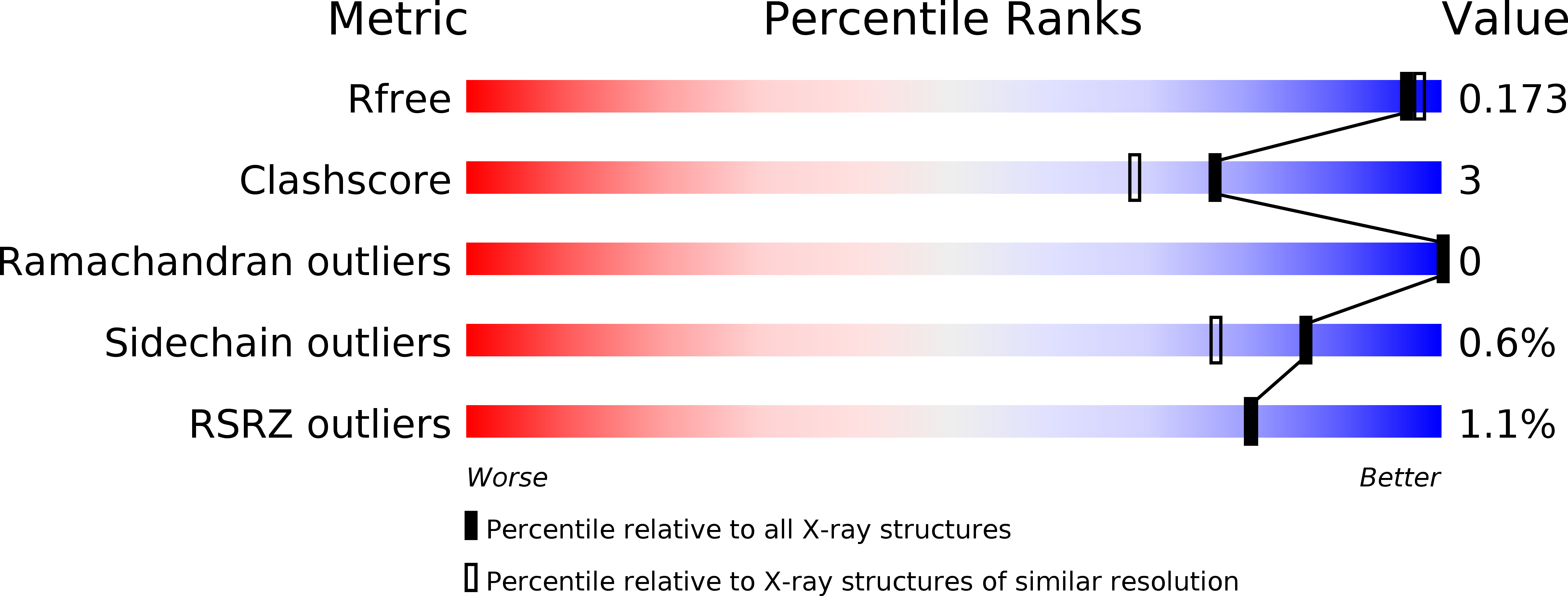
4U7A - PubMed Abstract:
Erb1 (Eukaryotic Ribosome Biogenesis 1) protein is essential for the maturation of the ribosomal 60S subunit. Functional studies in yeast and mammalian cells showed that altogether with Nop7 and Ytm1 it forms a stable subcomplex called PeBoW that is crucial for a correct rRNA processing. The exact function of the protein within the process remains unknown. The N-terminal region of the protein includes a well conserved region shown to be involved in PeBoW complex formation whereas the carboxy-terminal half was predicted to contain seven WD40 repeats. This first structural report on Erb1 from yeast describes the architecture of a seven-bladed β-propeller domain that revealed a characteristic extra motif formed by two α-helices and a β-strand that insert within the second WD repeat. We performed analysis of molecular surface and crystal packing, together with multiple sequence alignment and comparison of the structure with other β-propellers, in order to identify areas that are more likely to mediate protein-protein interactions. The abundance of many positively charged residues on the surface of the domain led us to investigate whether the propeller of Erb1 might be involved in RNA binding. Three independent assays confirmed that the protein interacted in vitro with polyuridilic acid (polyU), thus suggesting a possible role of the domain in rRNA rearrangement during ribosome biogenesis.
Organizational Affiliation:
Instituto de Biología Molecular y Celular, Universidad Miguel Hernández, Avda. del Ferrocarril s/n, 03202 Elche (Alicante), Spain; Instituto de Biocomputación y Física de los Sistemas Complejos (BIFI), 50009 Zaragoza, Spain.