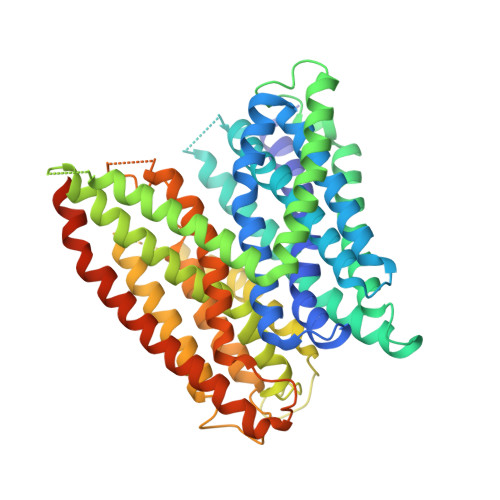
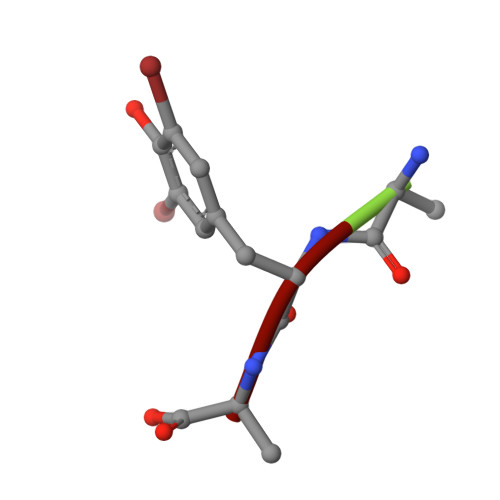
Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2.
Guettou, F., Quistgaard, E.M., Raba, M., Moberg, P., Low, C., Nordlund, P.(2014) Nat Struct Mol Biol 21: 728
- PubMed: 25064511
- DOI: https://doi.org/10.1038/nsmb.2860
- Primary Citation of Related Structures:
4TPG, 4TPH, 4TPJ - PubMed Abstract:
Peptide transporters of the PepT family have key roles in the transport of di- and tripeptides across membranes as well as in the absorption of orally administered drugs in the small intestine. We have determined structures of a PepT transporter from Shewanella oneidensis (PepT(So2)) in complex with three different peptides. The peptides bind in a large cavity lined by residues that are highly conserved in human PepT1 and PepT2. The bound peptides adopt extended conformations with their N termini clamped into a conserved polar pocket. A positively charged patch allows differential interactions with the C-terminal carboxylates of di- and tripeptides. Here we identify three pockets for peptide side chain interactions, and our binding studies define differential roles of these pockets for the recognition of different subtypes of peptide side chains.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden.