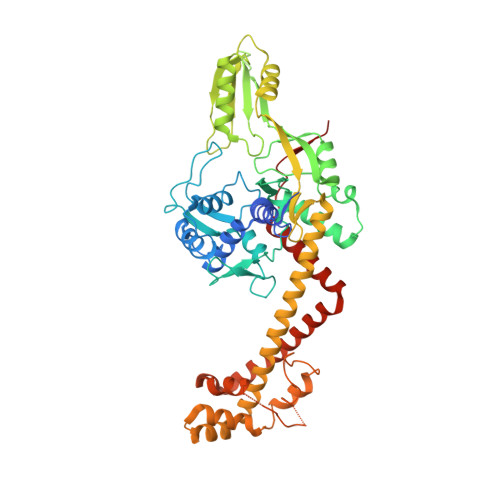
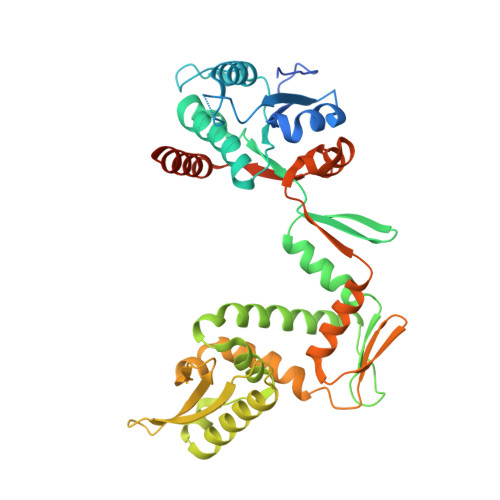
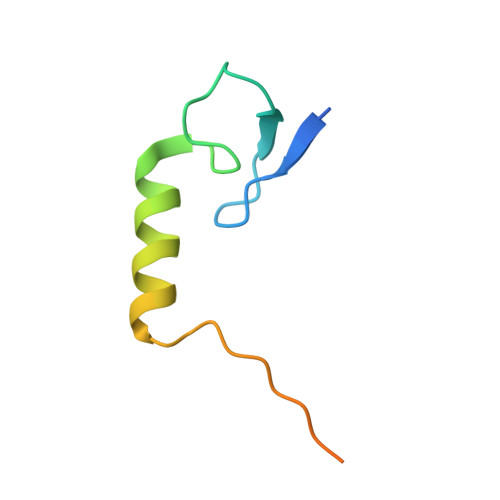
Direct control of type IIA topoisomerase activity by a chromosomally encoded regulatory protein.
Vos, S.M., Lyubimov, A.Y., Hershey, D.M., Schoeffler, A.J., Sengupta, S., Nagaraja, V., Berger, J.M.(2014) Genes Dev 28: 1485-1497
- PubMed: 24990966
- DOI: https://doi.org/10.1101/gad.241984.114
- Primary Citation of Related Structures:
4TMA - PubMed Abstract:
Precise control of supercoiling homeostasis is critical to DNA-dependent processes such as gene expression, replication, and damage response. Topoisomerases are central regulators of DNA supercoiling commonly thought to act independently in the recognition and modulation of chromosome superstructure; however, recent evidence has indicated that cells tightly regulate topoisomerase activity to support chromosome dynamics, transcriptional response, and replicative events. How topoisomerase control is executed and linked to the internal status of a cell is poorly understood. To investigate these connections, we determined the structure of Escherichia coli gyrase, a type IIA topoisomerase bound to YacG, a recently identified chromosomally encoded inhibitor protein. Phylogenetic analyses indicate that YacG is frequently associated with coenzyme A (CoA) production enzymes, linking the protein to metabolism and stress. The structure, along with supporting solution studies, shows that YacG represses gyrase by sterically occluding the principal DNA-binding site of the enzyme. Unexpectedly, YacG acts by both engaging two spatially segregated regions associated with small-molecule inhibitor interactions (fluoroquinolone antibiotics and the newly reported antagonist GSK299423) and remodeling the gyrase holoenzyme into an inactive, ATP-trapped configuration. This study establishes a new mechanism for the protein-based control of topoisomerases, an approach that may be used to alter supercoiling levels for responding to changes in cellular state.
Organizational Affiliation:
Department of Molecular and Cell Biology.