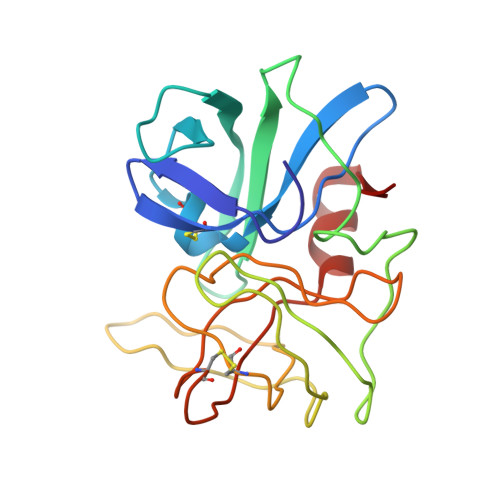
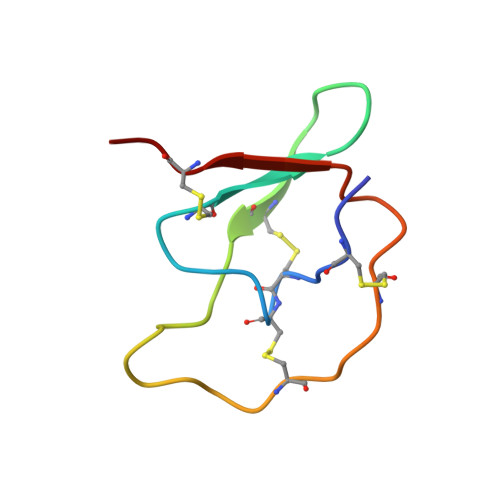
Structure of the complex of Streptomyces griseus proteinase B and polypeptide chymotrypsin inhibitor-1 from Russet Burbank potato tubers at 2.1 A resolution.
Greenblatt, H.M., Ryan, C.A., James, M.N.(1989) J Mol Biol 205: 201-228
- PubMed: 2494344
- DOI: https://doi.org/10.1016/0022-2836(89)90376-8
- Primary Citation of Related Structures:
4SGB - PubMed Abstract:
A low molecular weight protein inhibitor of serine proteinases from Russet Burbank potato tubers, polypeptide chymotrypsin inhibitor-1 (PCI-1), has been crystallized in complex with Streptomyces griseus proteinase B (SGPB). The three-dimensional structure of the complex has been solved at 2.1 A resolution by the molecular replacement method and has been refined to a final R-factor (= sigma[[Fo[-[Fc[[/sigma[Fo[) of 0.142 (8.0 to 2.1 A resolution data). The reactive site bond of PCI-1 (Leu38I to Asn39I) is intact in the complex, and there is no significant distortion of the peptide from planarity. The distance between the active site serine O gamma of SGPB and the carbonyl carbon of the scissile bond of PCI-1 is 2.8 A (1 A = 0.1 nm). The inhibitor has little secondary structure, having a three-stranded antiparallel beta-sheet on the side opposite the reactive site and four beta-turns. PCI-1 has four disulphide bridges; these presumably take the place of extensive secondary structure in keeping the reactive site conformationally constrained. The pairing of the cystine residues, which had not been characterized chemically, is as follows: Cys3I to Cys40I, Cys6I to Cys24I, Cys7I to Cys36I, and Cys13I to Cys49I. The molecular structure of SGPB in the PCI-1 complex agrees closely with the structure of SGPB complexed with the third domain of the turkey ovomucoid inhibitor (OMTKY3). A least-squares overlap of all atoms in SGPB gives a root-mean-square difference of 0.37 A. One of the loops of SGPB (Ser35 to Gly40) differs in conformation in the two complexes by more than 2.0 A root-mean-square for the main-chain atoms. Thr39 displays the largest differences with the carbonyl carbon atom deviating by 3.6 A. This conformational alternative is a result of the differences in the molecular structures of the P'4 residues following the reactive site bonds of the two inhibitors. This displacement avoids a close contact (1.3 A) between the carbonyl oxygen of Ser38 of SGPB and Pro42I C beta of PCI-1. The solvent structure of the PCI-1-SGPB complex includes 179 waters, two sulphate or phosphate ions, and one calcium or potassium ion, which appears to play a role in crystal formation. The molecular structure of PCI-1 determined here has allowed the proposal of a model for the structure of a two-domain inhibitor from potatoes and tomatoes, inhibitor II.(ABSTRACT TRUNCATED AT 400 WORDS)
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton.