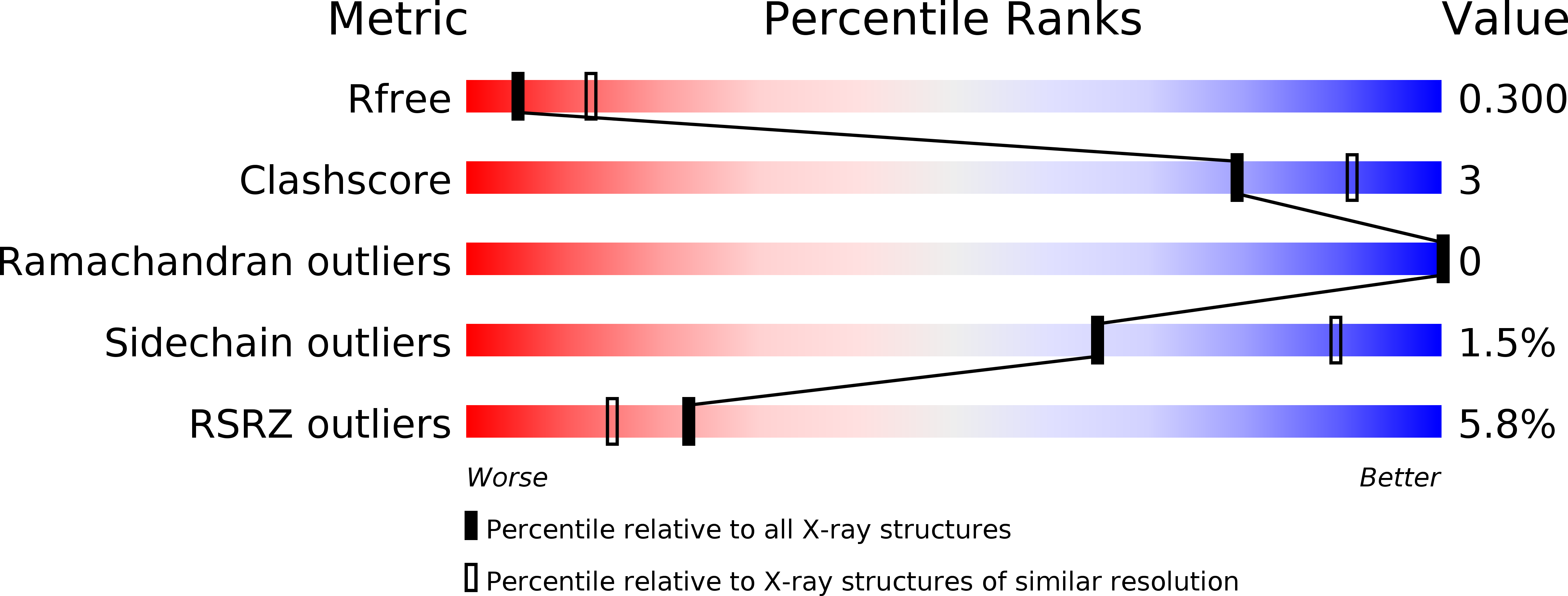
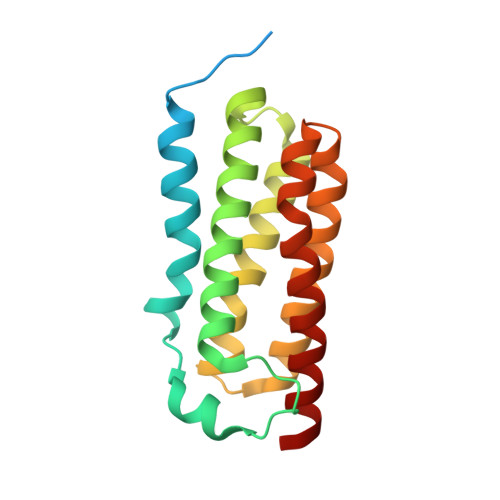
Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Guo, Y., Kalathur, R.C., Liu, Q., Kloss, B., Bruni, R., Ginter, C., Kloppmann, E., Rost, B., Hendrickson, W.A.(2015) Science 347: 551-555
- PubMed: 25635100
- DOI: https://doi.org/10.1126/science.aaa1534
- Primary Citation of Related Structures:
4RYI, 4RYJ, 4RYM, 4RYN, 4RYO, 4RYQ, 4RYR - PubMed Abstract:
Translocator proteins (TSPOs) bind steroids and porphyrins, and they are implicated in many human diseases, for which they serve as biomarkers and therapeutic targets. TSPOs have tryptophan-rich sequences that are highly conserved from bacteria to mammals. Here we report crystal structures for Bacillus cereus TSPO (BcTSPO) down to 1.7 Å resolution, including a complex with the benzodiazepine-like inhibitor PK11195. We also describe BcTSPO-mediated protoporphyrin IX (PpIX) reactions, including catalytic degradation to a previously undescribed heme derivative. We used structure-inspired mutations to investigate reaction mechanisms, and we showed that TSPOs from Xenopus and man have similar PpIX-directed activities. Although TSPOs have been regarded as transporters, the catalytic activity in PpIX degradation suggests physiological importance for TSPOs in protection against oxidative stress.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA.