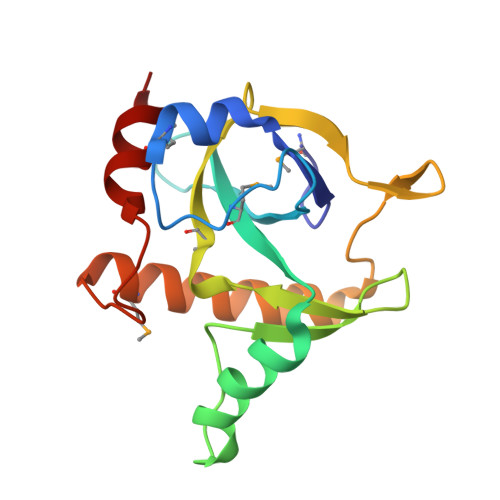
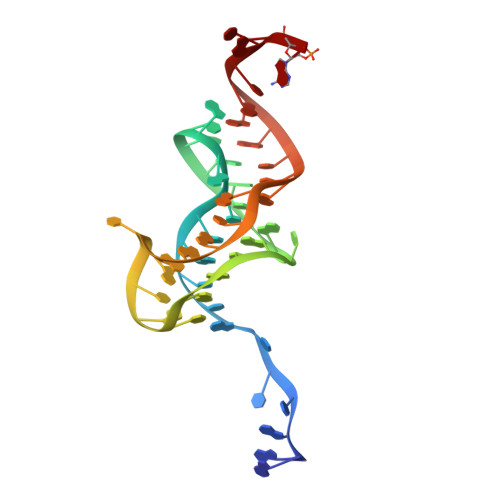
Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein-RNA toxin-antitoxin system.
Rao, F., Short, F.L., Voss, J.E., Blower, T.R., Orme, A.L., Whittaker, T.E., Luisi, B.F., Salmond, G.P.(2015) Nucleic Acids Res 43: 9529-9540
- PubMed: 26350213
- DOI: https://doi.org/10.1093/nar/gkv868
- Primary Citation of Related Structures:
4RMO - PubMed Abstract:
Genes encoding toxin-antitoxin (TA) systems are near ubiquitous in bacterial genomes and they play key roles in important aspects of bacterial physiology, including genomic stability, formation of persister cells under antibiotic stress, and resistance to phage infection. The CptIN locus from Eubacterium rectale is a member of the recently-discovered Type III class of TA systems, defined by a protein toxin suppressed by direct interaction with a structured RNA antitoxin. Here, we present the crystal structure of the CptIN protein-RNA complex to 2.2 Å resolution. The structure reveals a new heterotetrameric quaternary organization for the Type III TA class, and the RNA antitoxin bears a novel structural feature of an extended A-twist motif within the pseudoknot fold. The retention of a conserved ribonuclease active site as well as traits normally associated with TA systems, such as plasmid maintenance, implicates a wider functional role for Type III TA systems. We present evidence for the co-variation of the Type III component pair, highlighting a distinctive evolutionary process in which an enzyme and its substrate co-evolve.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge CB2 1QW, UK.